The Human Centrosomal Protein CCDC146 Binds Chlamydia trachomatis Inclusion Membrane Protein CT288 and Is Recruited to the Periphery of the Chlamydia-Containing Vacuole
- PMID: 30094225
- PMCID: PMC6070772
- DOI: 10.3389/fcimb.2018.00254
The Human Centrosomal Protein CCDC146 Binds Chlamydia trachomatis Inclusion Membrane Protein CT288 and Is Recruited to the Periphery of the Chlamydia-Containing Vacuole
Abstract
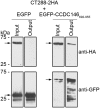
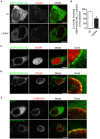
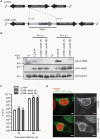
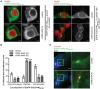
Chlamydia trachomatis is an obligate intracellular human pathogen causing mainly ocular and genital infections of significant clinical and public health impact. C. trachomatis multiplies intracellularly in a membrane bound vacuole, known as inclusion. Both extracellularly and from within the inclusion, C. trachomatis uses a type III secretion system to deliver several effector proteins into the cytoplasm of host cells. A large proportion of these effectors, the inclusion membrane (Inc) proteins, are exposed to the host cell cytosol but possess a characteristic hydrophobic domain mediating their insertion in the inclusion membrane. By yeast two-hybrid, we found that C. trachomatis Inc CT288 interacts with the human centrosomal protein CCDC146 (coiled-coil domain-containing protein 146). The interaction was also detected by co-immunoprecipitation in mammalian cells either ectopically expressing CCDC146 and CT288 or ectopically expressing CCDC146 and infected by a C. trachomatis strain expressing epitope-tagged and inclusion membrane-localized CT288. In uninfected mammalian cells, ectopically expressed full-length CCDC146 (955 amino acid residues) localized at the centrosome; but in cells infected by wild-type C. trachomatis, its centrosomal localization was less evident and CCDC146 accumulated around the inclusion. Recruitment of CCDC146 to the inclusion periphery did not require intact host Golgi, microtubules or microfilaments, but was dependent on chlamydial protein synthesis. Full-length CCDC146 also accumulated at the periphery of the inclusion in cells infected by a C. trachomatis ct288 mutant; however, a C-terminal fragment of CCDC146 (residues 692-955), which interacts with CT288, showed differences in localization at the periphery of the inclusion in cells infected by wild-type or ct288 mutant C. trachomatis. This suggests a model in which chlamydial proteins other than CT288 recruit CCDC146 to the periphery of the inclusion, where the CT288-CCDC146 interaction might contribute to modulate the function of this host protein.
Keywords: Chlamydia trachomatis; Inc proteins; centrosome; host-pathogen interactions; type III secretion.
Figures


References
-
- Aeberhard L., Banhart S., Fischer M., Jehmlich N., Rose L., Koch S., et al. . (2015). The proteome of the isolated Chlamydia trachomatis containing vacuole reveals a complex trafficking platform enriched for retromer components. PLoS Pathog. 11:e1004883. 10.1371/journal.ppat.1004883 - DOI - PMC - PubMed
-
- Almeida F., Borges V., Ferreira R., Borrego M. J., Gomes J. P., Mota L. J. (2012). Polymorphisms in inc proteins and differential expression of inc genes among Chlamydia trachomatis Strains correlate with invasiveness and tropism of lymphogranuloma venereum isolates. J. Bacteriol. 194, 6574–6585. 10.1128/JB.01428-12 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

