Comparison of 2-Aminobenzamide, Procainamide and Rapi Fluor-MS as Derivatizing Agents for High-Throughput HILIC-UPLC-FLR-MS N-glycan Analysis
- PMID: 30094234
- PMCID: PMC6070730
- DOI: 10.3389/fchem.2018.00324
Comparison of 2-Aminobenzamide, Procainamide and Rapi Fluor-MS as Derivatizing Agents for High-Throughput HILIC-UPLC-FLR-MS N-glycan Analysis
Abstract
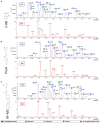
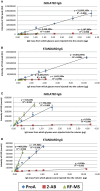
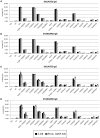
Rising awareness of the universal importance of protein N-glycosylation governs the development of further advances in N-glycan analysis. Nowadays it is well known that correct glycosylation is essential for proper protein function, which emanates from its important role in many physiological processes. Furthermore, glycosylation is involved in pathophysiology of multiple common complex diseases. In the vast majority of cases, N-glycosylation profiles are analyzed from enzymatically released glycans, which can be further derivatized in order to enhance the sensitivity of the analysis. Techniques wherein derivatized N-glycans are profiled using hydrophilic interaction chromatography (HILIC) with fluorescence (FLR) and mass spectrometry (MS) detection are now routinely performed in a high-throughput manner. Therefore, we aimed to examine the performance of frequently used labeling compounds -2-aminiobenzamide (2-AB) and procainamide (ProA), and the recently introduced RapiFluor-MS (RF-MS) fluorescent tag. In all experiments N-glycans were released by PNGase F, fluorescently derivatized, purified by HILIC solid phase extraction and profiled using HILIC-UPLC-FLR-MS. We assessed sensitivity, linear range, limit of quantification (LOQ), repeatability and labeling efficiency for all three labels. For this purpose, we employed in-house prepared IgG and a commercially available IgG as a model glycoprotein. All samples were analyzed in triplicates using different amounts of starting material. We also tested the performance of all three labels in a high-throughput setting on 68 different IgG samples, all in duplicates and 22 identical IgG standards. In general, ProA labeled glycans had the highest FLR sensitivity (15-fold and 4-fold higher signal intensities compared to 2-AB and RF-MS respectively) and RF-MS had the highest MS sensitivity (68-fold and 2-fold higher signal intensities compared to 2-AB and ProA, respectively). ProA and RF-MS showed comparable limits of quantification with both FLR and MS detection, whilst 2-AB exhibited the lowest sensitivity. All labeling procedures showed good and comparable repeatability. Furthermore, the results indicated that labeling efficiency was very similar for all three labels. In conclusion, all three labels are a good choice for N-glycan derivatization in high-throughput HILIC-UPLC-FLR-MS N-glycan analysis, although ProA and RF-MS are a better option when higher sensitivity is needed.
Keywords: 2-aminobenzamide; HILIC; IgG; N-glycans; RapiFluor-MS; fluorescence; mass spectrometry; procainamide.
Figures
References
-
- Beck A. (ed.). (2013). Glycosylation Engineering of Biopharmaceuticals. 1st Edn. New York, NY: Hum.
LinkOut - more resources
Full Text Sources
Other Literature Sources

