Coral microbiome database: Integration of sequences reveals high diversity and relatedness of coral-associated microbes
- PMID: 30094953
- PMCID: PMC7379671
- DOI: 10.1111/1758-2229.12686
Coral microbiome database: Integration of sequences reveals high diversity and relatedness of coral-associated microbes
Abstract
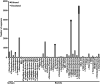
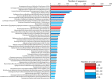
Coral-associated microorganisms are thought to play a fundamental role in the health and ecology of corals, but understanding of specific coral-microbial interactions are lacking. In order to create a framework to examine coral-microbial specificity, we integrated and phylogenetically compared 21,100 SSU rRNA gene Sanger-produced sequences from bacteria and archaea associated with corals from previous studies, and accompanying host, location and publication metadata, to produce the Coral Microbiome Database. From this database, we identified 39 described and candidate phyla of Bacteria and two Archaea phyla associated with corals, demonstrating that corals are one of the most phylogenetically diverse animal microbiomes. Secondly, this new phylogenetic resource shows that certain microorganisms are indeed specific to corals, including evolutionary distinct hosts. Specifically, we identified 2-37 putative monophyletic, coral-specific sequence clusters within bacterial genera associated with the greatest number of coral species (Vibrio, Endozoicomonas and Ruegeria) as well as functionally relevant microbial taxa ("Candidatus Amoebophilus", "Candidatus Nitrosopumilus" and under recognized cyanobacteria). This phylogenetic resource provides a framework for more targeted studies of corals and their specific microbial associates, which is timely given the escalated need to understand the role of the coral microbiome and its adaptability to changing ocean and reef conditions.
© 2018 The Authors. Environmental Microbiology Reports published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures






References
-
- Ainsworth, T.D. , Fordyce, A.J. , and Camp, E.F. (2017) The other microeukaryotes of the coral reef microbiome. Trends Microbiol 25: 980–991. - PubMed
-
- Apprill, A. , Hughen, K. , and Mincer, T.J. (2013) Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ Microbiol 15: 2063–2072. - PubMed
-
- Barott, K.L. , Rodriguez‐Brito, B. , Janouškovec, J. , Marhaver, K.L. , Smith, J.E. , Keeling, P. , and Rohwer, F.L. (2011) Microbial diversity associated with four functional groups of benthic reef algae and the reef‐building coral Montastraea annularis . Environ Microbiol 13: 1192–1204. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

