Characterization of the two conformations adopted by the T3SS inner-membrane protein PrgK
- PMID: 30095200
- PMCID: PMC6194235
- DOI: 10.1002/pro.3447
Characterization of the two conformations adopted by the T3SS inner-membrane protein PrgK
Abstract
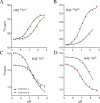
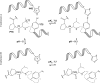
The pathogenic bacterium Salmonella enterica serovar Typhimurium utilizes two type III secretion systems (T3SS) to inject effector proteins into target cells upon infection. The T3SS secretion apparatus (the injectisome) is a large macromolecular assembly composed of over twenty proteins, many in highly oligomeric states. A sub-structure of the injectisome, termed the basal body, spans both membranes and the periplasmic space of the bacterium. It is primarily composed of three integral membranes proteins, InvG, PrgH, and PrgK, that form ring structures through which components are secreted. In particular, PrgK possesses a periplasmic region consisting of two globular domains joined by a linker polypeptide. We showed previously that in isolation, this region adopts two distinct conformations, of with only one is observed in the assembled basal body complex. Here, using NMR spectroscopy, we further characterize these two conformations. In particular, we demonstrate that the interaction of the linker region with the first globular domain, as found in the intact basal body, is dependent upon the cis conformation of the Leu77-Pro78 peptide. Furthermore, this interaction is pH-dependent due to coupling with hydrogen bond formation between Tyr75 and His42 in its neutral Nδ1 H tautomeric form. This pH-dependent interaction may play a role in the regulation of the secretion apparatus disassembly in the context of bacterial infection.
Keywords: NMR; Salmonella enterica serovar Typhimurium; bacterial secretion systems; protein dynamics.
© 2018 The Protein Society.
Figures





References
-
- Costa TR, Felisberto‐Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G (2015) Secretion systems in Gram‐negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. - PubMed
-
- Burkinshaw BJ, Strynadka NC (2014) Assembly and structure of the T3SS. Biochim Biophys Acta 1843:1649–1663. - PubMed
-
- Moest TP, Meresse S (2013) Salmonella T3SSs: successful mission of the secret(ion) agents. Curr Opin Microbiol 16:38–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

