Atlas of the Radical SAM Superfamily: Divergent Evolution of Function Using a "Plug and Play" Domain
- PMID: 30097089
- PMCID: PMC6445391
- DOI: 10.1016/bs.mie.2018.06.004
Atlas of the Radical SAM Superfamily: Divergent Evolution of Function Using a "Plug and Play" Domain
Abstract
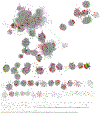
The radical SAM superfamily contains over 100,000 homologous enzymes that catalyze a remarkably broad range of reactions required for life, including metabolism, nucleic acid modification, and biogenesis of cofactors. While the highly conserved SAM-binding motif responsible for formation of the key 5'-deoxyadenosyl radical intermediate is a key structural feature that simplifies identification of superfamily members, our understanding of their structure-function relationships is complicated by the modular nature of their structures, which exhibit varied and complex domain architectures. To gain new insight about these relationships, we classified the entire set of sequences into similarity-based subgroups that could be visualized using sequence similarity networks. This superfamily-wide analysis reveals important features that had not previously been appreciated from studies focused on one or a few members. Functional information mapped to the networks indicates which members have been experimentally or structurally characterized, their known reaction types, and their phylogenetic distribution. Despite the biological importance of radical SAM chemistry, the vast majority of superfamily members have never been experimentally characterized in any way, suggesting that many new reactions remain to be discovered. In addition to 20 subgroups with at least one known function, we identified additional subgroups made up entirely of sequences of unknown function. Importantly, our results indicate that even general reaction types fail to track well with our sequence similarity-based subgroupings, raising major challenges for function prediction for currently identified and new members that continue to be discovered. Interactive similarity networks and other data from this analysis are available from the Structure-Function Linkage Database.
Keywords: Classification of Radical SAM enzymes by sequence similarity; Multiple domain structures of radical SAM superfamily enzymes; Phylogenetic representation; Radical SAM superfamily census; Sequence similarity networks; Structure–function mapping; Subgroups and families in the radical SAM superfamily.
© 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS STATEMENT
None
Figures




References
-
- Anantharaman V, Koonin EV, & Aravind L (2001). TRAM, a predicted RNA-binding domain, common to tRNA uracil methylation and adenine thiolation enzymes. FEMS Microbiol Lett, 197(2), 215–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

