Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion
- PMID: 30097514
- PMCID: PMC6168255
- DOI: 10.1083/jcb.201804028
Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion
Abstract
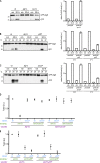
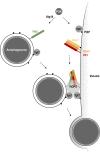
Autophagy mediates the bulk degradation of cytoplasmic material, particularly during starvation. Upon the induction of autophagy, autophagosomes form a sealed membrane around cargo, fuse with a lytic compartment, and release the cargo for degradation. The mechanism of autophagosome-vacuole fusion is poorly understood, although factors that mediate other cellular fusion events have been implicated. In this study, we developed an in vitro reconstitution assay that enables systematic discovery and dissection of the players involved in autophagosome-vacuole fusion. We found that this process requires the Atg14-Vps34 complex to generate PI3P and thus recruit the Ypt7 module to autophagosomes. The HOPS-tethering complex, recruited by Ypt7, is required to prepare SNARE proteins for fusion. Furthermore, we discovered that fusion requires the R-SNARE Ykt6 on the autophagosome, together with the Q-SNAREs Vam3, Vam7, and Vti1 on the vacuole. These findings shed new light on the mechanism of autophagosome-vacuole fusion and reveal that the R-SNARE Ykt6 is required for this process.
© 2018 Bas et al.
Figures
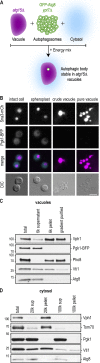
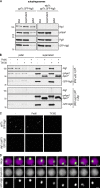
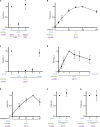

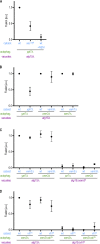
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

