Glycomics@ExPASy: Bridging the Gap
- PMID: 30097532
- PMCID: PMC6210229
- DOI: 10.1074/mcp.RA118.000799
Glycomics@ExPASy: Bridging the Gap
Abstract
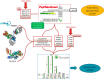
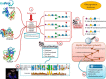
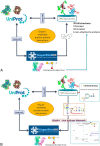
Glycomics@ExPASy (https://www.expasy.org/glycomics) is the glycomics tab of ExPASy, the server of SIB Swiss Institute of Bioinformatics. It was created in 2016 to centralize web-based glycoinformatics resources developed within an international network of glycoscientists. The hosted collection currently includes mainly databases and tools created and maintained at SIB but also links to a range of reference resources popular in the glycomics community. The philosophy of our toolbox is that it should be {glycoscientist AND protein scientist}-friendly with the aim of (1) popularizing the use of bioinformatics in glycobiology and (2) emphasizing the relationship between glycobiology and protein-oriented bioinformatics resources. The scarcity of data bridging these two disciplines led us to design tools as interactive as possible based on database connectivity to facilitate data exploration and support hypothesis building. Glycomics@ExPASy was designed, and is developed, with a long-term vision in close collaboration with glycoscientists to meet as closely as possible the growing needs of the community for glycoinformatics.
Keywords: Bioinformatics; Bioinformatics software; Glycomics; Glycoproteomics; Glycosylation.
© 2018 Mariethoz et al.
Figures


References
-
- National Research Council (US) Committee on Assessing the Importance and Impact of Glycomics and Glycosciences. (2012) Transforming Glycoscience: A Roadmap for the Future (National Academies Press (US), Washington (DC)) - PubMed
-
- Lundborg M., and Widmalm G. (2011) Structural analysis of glycans by NMR chemical shift prediction. Anal. Chem. 83, 1514–1517 - PubMed
-
- Adamczyk B., Stöckmann H., O'Flaherty R., Karlsson N. G., and Rudd P. M. (2017) in High-Throughput Glycomics and Glycoproteomics, eds Lauc G, Wuhrer M (Springer New York, New York, NY: ), pp 97–108
-
- Ruhaak L. R., Hennig R., Huhn C., Borowiak M., Dolhain R. J. E. M., Deelder A. M., Rapp E., and Wuhrer M. (2010) Optimized workflow for preparation of APTS-labeled N-glycans allowing high-throughput analysis of human plasma glycomes using 48-channel multiplexed CGE-LIF. J. Proteome Res. 9, 6655–6664 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

