Flavin oxidation in flavin-dependent N-monooxygenases
- PMID: 30098072
- PMCID: PMC6295899
- DOI: 10.1002/pro.3487
Flavin oxidation in flavin-dependent N-monooxygenases
Abstract
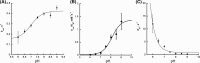
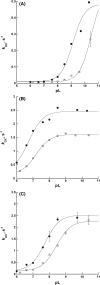
Siderophore A (SidA) from Aspergillus fumigatus is a flavin-containing monooxygenase that hydroxylates ornithine (Orn) at the amino group of the side chain. Lysine (Lys) also binds to the active site of SidA; however, hydroxylation is not efficient and H2 O2 is the main product. The effect of pH on steady-state kinetic parameters was measured and the results were consistent with Orn binding with the side chain amino group in the neutral form. From the pH dependence on flavin oxidation in the absence of Orn, a pKa value >9 was determined and assigned to the FAD-N5 atom. In the presence of Orn, the pH dependence displayed a pKa value of 6.7 ±0.1 and of 7.70 ±0.10 in the presence of Lys. Q102 interacts with NADPH and, upon mutation to alanine, leads to destabilization of the C4a-hydroperoxyflavin (FADOOH ). Flavin oxidation with Q102A showed a pKa value of ~8.0. The data are consistent with the pKa of the FAD N5-atom being modulated to a value >9 in the absence of Orn, which aids in the stabilization of FADOOH . Changes in the FAD-N5 environment lead to a decrease in the pKa value, which facilitates elimination of H2 O2 or H2 O. These findings are supported by solvent kinetic isotope effect experiments, which show that proton transfer from the FAD N5-atom is rate limiting in the absence of a substrate, however, is significantly less rate limiting in the presence of Orn and or Lys.
Keywords: flavin-dependent monooxygneases; hydroperoxyflavin; ornithine hydroxylase; oxidation; pH profile; siderophore; solvent kinetic isotope effect.
© 2018 The Protein Society.
Figures




Similar articles
-
Trapping conformational states of a flavin-dependent N-monooxygenase in crystallo reveals protein and flavin dynamics.J Biol Chem. 2020 Sep 18;295(38):13239-13249. doi: 10.1074/jbc.RA120.014750. Epub 2020 Jul 28. J Biol Chem. 2020. PMID: 32723870 Free PMC article.
-
Structural Determinants of Flavin Dynamics in a Class B Monooxygenase.Biochemistry. 2020 Dec 8;59(48):4609-4616. doi: 10.1021/acs.biochem.0c00783. Epub 2020 Nov 23. Biochemistry. 2020. PMID: 33226785
-
Arg279 is the key regulator of coenzyme selectivity in the flavin-dependent ornithine monooxygenase SidA.Biochim Biophys Acta. 2014 Apr;1844(4):778-84. doi: 10.1016/j.bbapap.2014.02.005. Epub 2014 Feb 15. Biochim Biophys Acta. 2014. PMID: 24534646
-
Mechanistic and structural studies of the N-hydroxylating flavoprotein monooxygenases.Bioorg Chem. 2011 Dec;39(5-6):171-7. doi: 10.1016/j.bioorg.2011.07.006. Epub 2011 Aug 5. Bioorg Chem. 2011. PMID: 21871647 Free PMC article. Review.
-
Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases.Biochem Biophys Res Commun. 2005 Dec 9;338(1):590-8. doi: 10.1016/j.bbrc.2005.09.081. Epub 2005 Sep 26. Biochem Biophys Res Commun. 2005. PMID: 16236251 Review.
Cited by
-
Trapping conformational states of a flavin-dependent N-monooxygenase in crystallo reveals protein and flavin dynamics.J Biol Chem. 2020 Sep 18;295(38):13239-13249. doi: 10.1074/jbc.RA120.014750. Epub 2020 Jul 28. J Biol Chem. 2020. PMID: 32723870 Free PMC article.
-
Substrate Electronics Dominate the Rate of Reductive Dehalogenation Promoted by the Flavin-Dependent Iodotyrosine Deiodinase.Biochemistry. 2023 Apr 4;62(7):1298-1306. doi: 10.1021/acs.biochem.3c00041. Epub 2023 Mar 9. Biochemistry. 2023. PMID: 36892456 Free PMC article.
-
Photoinduced Covalent Irreversible Inactivation of Proline Dehydrogenase by S-Heterocycles.ACS Chem Biol. 2021 Nov 19;16(11):2268-2279. doi: 10.1021/acschembio.1c00427. Epub 2021 Sep 20. ACS Chem Biol. 2021. PMID: 34542291 Free PMC article.
-
A biosynthetic aspartate N-hydroxylase performs successive oxidations by holding intermediates at a site away from the catalytic center.J Biol Chem. 2023 Jul;299(7):104904. doi: 10.1016/j.jbc.2023.104904. Epub 2023 Jun 10. J Biol Chem. 2023. PMID: 37302552 Free PMC article.
References
-
- Fischbach MA, Lin H, Liu DR, Walsh CT (2006) How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol 2:132–138. - PubMed
-
- Esuola CO, Babaloa OO, Heine T, Schwabe R, Schlomann M, Tischler D (2016) Identification and characterization of a FAD‐dependent putrescine N‐hydroxylase (GorA) from Gordonia rubripertincta CWB2. J Mol Cataly B 134:378–389.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

