The NETest: The Clinical Utility of Multigene Blood Analysis in the Diagnosis and Management of Neuroendocrine Tumors
- PMID: 30098712
- PMCID: PMC6716518
- DOI: 10.1016/j.ecl.2018.05.002
The NETest: The Clinical Utility of Multigene Blood Analysis in the Diagnosis and Management of Neuroendocrine Tumors
Abstract
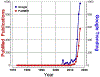
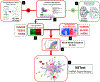
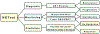
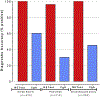
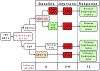
The neuroendocrine neoplasms test (NETest) is a multianalyte liquid biopsy that measures neuroendocrine tumor gene expression in blood. This unique signature precisely defines the biological activity of an individual tumor in real time. The assay meets the 3 critical requirements of an optimal biomarker: diagnostic accuracy, prognostic value, and predictive therapeutic assessment. NETest performance metrics are sensitivity and specificity and in head-to-head comparison are 4-fold to 10-fold more accurate than chromogranin A. NETest accurately identifies completeness of surgery and response to somatostatin analogs. Clinical registry data demonstrate significant clinical utility in watch/wait programs.
Keywords: Biomarker; Blood; Bronchopulomary carcinoid; Multigene blood analysis; NETest; Neuroendocrine tumors; PCR; Peptide receptor radionuclide therapy; Progression; Transcript.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

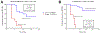
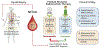
References
-
- de Mestier L, Dromain C, d’Assignies G, et al. Evaluating digestive neuroendocrine tumor progression and therapeutic responses in the era of targeted therapies: state of the art. Endocr Relat Cancer 2014;21(3):R105–20. Print 2014. - PubMed
-
- Bergsland EK. The evolving landscape of neuroendocrine tumors. Semin Oncol 2013;40(1):4–22. - PubMed
-
- Wang H, Chen Y, Fernandez-Del Castillo C, et al. Heterogeneity in signaling pathways of gastroenteropancreatic neuroendocrine tumors: a critical look at notch signaling pathway. Mod Pathol 2012;24(10):143. - PubMed
-
- Sundin A, Rockall A. Therapeutic monitoring of gastroenteropancreatic neuroendocrine tumors: the challenges ahead. Neuroendocrinology 2012;96(4):261–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

