B-BOX DOMAIN PROTEIN28 Negatively Regulates Photomorphogenesis by Repressing the Activity of Transcription Factor HY5 and Undergoes COP1-Mediated Degradation
- PMID: 30099385
- PMCID: PMC6181009
- DOI: 10.1105/tpc.18.00226
B-BOX DOMAIN PROTEIN28 Negatively Regulates Photomorphogenesis by Repressing the Activity of Transcription Factor HY5 and Undergoes COP1-Mediated Degradation
Abstract
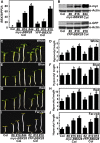
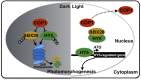
Plants have evolved a delicate molecular system to fine-tune their growth and development in response to dynamically changing light environments. In this study, we found that BBX28, a B-box domain protein, negatively regulates photomorphogenic development in a dose-dependent manner in Arabidopsis thaliana BBX28 interferes with the binding of transcription factor HY5 to the promoters of its target genes through physical interactions, thereby repressing its activity and negatively affecting HY5-regulated gene expression. In darkness, BBX28 associates with CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) and undergoes COP1-mediated degradation via the 26S proteasome system. Collectively, these results demonstrate that BBX28 acts as a key factor in the COP1-HY5 regulatory hub by maintaining proper HY5 activity to ensure normal photomorphogenic development in plants.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures







References
-
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. - PubMed
-
- Chang C.S., Li Y.H., Chen L.T., Chen W.C., Hsieh W.P., Shin J., Jane W.N., Chou S.J., Choi G., Hu J.M., Somerville S., Wu S.H. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54: 205–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

