Progress on Identifying and Characterizing the Human Proteome: 2018 Metrics from the HUPO Human Proteome Project
- PMID: 30099871
- PMCID: PMC6387656
- DOI: 10.1021/acs.jproteome.8b00441
Progress on Identifying and Characterizing the Human Proteome: 2018 Metrics from the HUPO Human Proteome Project
Abstract
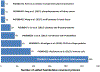
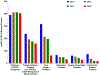
The Human Proteome Project (HPP) annually reports on progress throughout the field in credibly identifying and characterizing the human protein parts list and making proteomics an integral part of multiomics studies in medicine and the life sciences. NeXtProt release 2018-01-17, the baseline for this sixth annual HPP special issue of the Journal of Proteome Research, contains 17 470 PE1 proteins, 89% of all neXtProt predicted PE1-4 proteins, up from 17 008 in release 2017-01-23 and 13 975 in release 2012-02-24. Conversely, the number of neXtProt PE2,3,4 missing proteins has been reduced from 2949 to 2579 to 2186 over the past two years. Of the PE1 proteins, 16 092 are based on mass spectrometry results, and 1378 on other kinds of protein studies, notably protein-protein interaction findings. PeptideAtlas has 15 798 canonical proteins, up 625 over the past year, including 269 from SUMOylation studies. The largest reason for missing proteins is low abundance. Meanwhile, the Human Protein Atlas has released its Cell Atlas, Pathology Atlas, and updated Tissue Atlas, and is applying recommendations from the International Working Group on Antibody Validation. Finally, there is progress using the quantitative multiplex organ-specific popular proteins targeted proteomics approach in various disease categories.
Keywords: Biology and Disease-driven HPP (B/D-HPP); Chromosome-centric HPP (C-HPP); HPP Guidelines; Human Proteome Organization (HUPO); Human Proteome Project (HPP); PeptideAtlas; metrics; missing proteins; neXtProt.
Figures

References
-
- Legrain P; Aebersold R; Archakov A; Bairoch A; Bala K; Beretta L; Bergeron J; Borchers CH; Corthals GL; Costello CE; Deutsch EW; Domon B; Hancock W; He F; Hochstrasser D; Marko-Varga G; Salekdeh GH; Sechi S; Snyder M; Srivastava S; Uhlen M; Wu CH; Yamamoto T; Paik YK; Omenn GS, The Human Proteome Project: current state and future direction. Mol Cell Proteomics 2011, 10, (7), M111 009993. - PMC - PubMed
-
- Marko-Varga G; Omenn GS; Paik YK; Hancock WS, A first step toward completion of a genome-wide characterization of the human proteome. J. Proteome Res 2013, 12, (1), 1–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

