Tbx6 Induces Nascent Mesoderm from Pluripotent Stem Cells and Temporally Controls Cardiac versus Somite Lineage Diversification
- PMID: 30100166
- PMCID: PMC6190602
- DOI: 10.1016/j.stem.2018.07.001
Tbx6 Induces Nascent Mesoderm from Pluripotent Stem Cells and Temporally Controls Cardiac versus Somite Lineage Diversification
Abstract
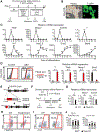
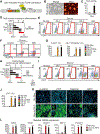
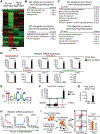
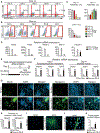
The mesoderm arises from pluripotent epiblasts and differentiates into multiple lineages; however, the underlying molecular mechanisms are unclear. Tbx6 is enriched in the paraxial mesoderm and is implicated in somite formation, but its function in other mesoderms remains elusive. Here, using direct reprogramming-based screening, single-cell RNA-seq in mouse embryos, and directed cardiac differentiation in pluripotent stem cells (PSCs), we demonstrated that Tbx6 induces nascent mesoderm from PSCs and determines cardiovascular and somite lineage specification via its temporal expression. Tbx6 knockout in mouse PSCs using CRISPR/Cas9 technology inhibited mesoderm and cardiovascular differentiation, whereas transient Tbx6 expression induced mesoderm and cardiovascular specification from mouse and human PSCs via direct upregulation of Mesp1, repression of Sox2, and activation of BMP/Nodal/Wnt signaling. Notably, prolonged Tbx6 expression suppressed cardiac differentiation and induced somite lineages, including skeletal muscle and chondrocytes. Thus, Tbx6 is critical for mesoderm induction and subsequent lineage diversification.
Keywords: Tbx6; cardiovascular; chondrocyte; mesoderm; pluripotent stem cell; skeletal muscle.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
T.S., M. Isomi, N.G., and M. Ieda are inventors on patent applications describing the Tbx6-induced reprogramming and differentiation into cardiac progenitors and cardiomyocytes.
Figures



Similar articles
-
Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells.Nature. 2011 Feb 17;470(7334):394-8. doi: 10.1038/nature09729. Nature. 2011. PMID: 21331042 Free PMC article.
-
Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6.Nature. 1998 Feb 12;391(6668):695-7. doi: 10.1038/35624. Nature. 1998. PMID: 9490412
-
Defective somite patterning in mouse embryos with reduced levels of Tbx6.Development. 2003 Apr;130(8):1681-90. doi: 10.1242/dev.00367. Development. 2003. PMID: 12620991
-
Myogenic progenitor specification from pluripotent stem cells.Semin Cell Dev Biol. 2017 Dec;72:87-98. doi: 10.1016/j.semcdb.2017.10.031. Semin Cell Dev Biol. 2017. PMID: 29107681 Free PMC article. Review.
-
Making muscle: skeletal myogenesis in vivo and in vitro.Development. 2017 Jun 15;144(12):2104-2122. doi: 10.1242/dev.151035. Development. 2017. PMID: 28634270 Review.
Cited by
-
Direct cell-fate conversion of somatic cells: Toward regenerative medicine and industries.Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(4):131-158. doi: 10.2183/pjab.96.012. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 32281550 Free PMC article. Review.
-
In silico screening system based on a transcription factors regulatory network only using transcriptomic data.PLoS One. 2025 Apr 7;20(4):e0319971. doi: 10.1371/journal.pone.0319971. eCollection 2025. PLoS One. 2025. PMID: 40193394 Free PMC article.
-
In the heart of the in vivo reprogramming.Stem Cell Investig. 2018 Oct 29;5:38. doi: 10.21037/sci.2018.10.03. eCollection 2018. Stem Cell Investig. 2018. PMID: 30498749 Free PMC article. No abstract available.
-
Single-cell transcriptomes in the heart: when every epigenome counts.Cardiovasc Res. 2023 Mar 17;119(1):64-78. doi: 10.1093/cvr/cvac040. Cardiovasc Res. 2023. PMID: 35325060 Free PMC article. Review.
-
Cooperation between ETS transcription factor ETV1 and histone demethylase JMJD1A in colorectal cancer.Int J Oncol. 2020 Dec;57(6):1319-1332. doi: 10.3892/ijo.2020.5133. Epub 2020 Oct 14. Int J Oncol. 2020. PMID: 33174020 Free PMC article.
References
-
- Chal J, and Pourquié O (2017). Making muscle: Skeletal myogenesis in vivo and in vitro. Development 144, 2104–2122. - PubMed
-
- Chal J, Oginuma M, Al Tanoury Z, Gobert B, Sumara O, Hick A, Bousson F, Zidouni Y, Mursch C, Moncuquet P, et al. (2015). Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol 33, 962–969. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

