Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex
- PMID: 30100186
- PMCID: PMC6108936
- DOI: 10.1016/j.cell.2018.07.020
Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex
Abstract
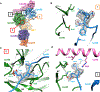
The methylation of histone 3 lysine 4 (H3K4) is carried out by an evolutionarily conserved family of methyltransferases referred to as complex of proteins associated with Set1 (COMPASS). The activity of the catalytic SET domain (su(var)3-9, enhancer-of-zeste, and trithorax) is endowed through forming a complex with a set of core proteins that are widely shared from yeast to humans. We obtained cryo-electron microscopy (cryo-EM) maps of the yeast Set1/COMPASS core complex at overall 4.0- to 4.4-Å resolution, providing insights into its structural organization and conformational dynamics. The Cps50 C-terminal tail weaves within the complex to provide a central scaffold for assembly. The SET domain, snugly positioned at the junction of the Y-shaped complex, is extensively contacted by Cps60 (Bre2), Cps50 (Swd1), and Cps30 (Swd3). The mobile SET-I motif of the SET domain is engaged by Cps30, explaining its key role in COMPASS catalytic activity toward higher H3K4 methylation states.
Keywords: COMPASS; MLL; cryo-EM; epigenetics; histone methylation.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing financial interests.
Figures


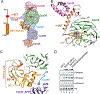


References
-
- Avdic V, Zhang P, Lanouette S, Groulx A, Tremblay V, Brunzelle J, and Couture JF (2011a). Structural and biochemical insights into MLL1 core complex assembly. Structure 19, 101–108. - PubMed
-
- Avdic V, Zhang P, Lanouette S, Voronova A, Skerjanc I, and Couture JF (2011b). Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic. FASEB J 25, 960–967. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, and Zhao K (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

