β-Defensin 1 Is Prominent in the Liver and Induced During Cholestasis by Bilirubin and Bile Acids via Farnesoid X Receptor and Constitutive Androstane Receptor
- PMID: 30100908
- PMCID: PMC6072844
- DOI: 10.3389/fimmu.2018.01735
β-Defensin 1 Is Prominent in the Liver and Induced During Cholestasis by Bilirubin and Bile Acids via Farnesoid X Receptor and Constitutive Androstane Receptor
Abstract
Background & aims: Knowledge about innate antimicrobial defense of the liver is limited. We investigated hepatic expression and regulation of antimicrobial peptides with focus on the human beta defensin-1 (hBD-1).
Methods: Radial diffusion assay was used to analyze antimicrobial activity of liver tissue. Different defensins including hBD-1 and its activator thioredoxin-1 (TXN) were analyzed in healthy and cholestatic liver samples by qPCR and immunostaining. Regulation of hBD-1 expression was studied in vitro and in vivo using bile duct-ligated mice. Regulation of hBD-1 via bilirubin and bile acids (BAs) was studied using siRNA.
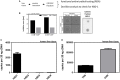
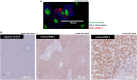
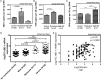
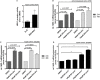
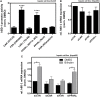
Results: We found strong antimicrobial activity of liver tissue against Escherichia coli. As a potential mediator of this antimicrobial activity we detected high expression of hBD-1 and TXN in hepatocytes, whereas other defensins were minimally expressed. Using a specific antibody for the reduced, antimicrobially active form of hBD-1 we found hBD-1 in co-localization with TXN within hepatocytes. hBD-1 was upregulated in cholestasis in a graded fashion. In cholestatic mice hepatic AMP expression (Defb-1 and Hamp) was enhanced. Bilirubin and BAs were able to induce hBD-1 in hepatic cell cultures in vitro. Treatment with siRNA and/or agonists demonstrated that the farnesoid X receptor (FXR) mediates basal expression of hBD-1, whereas both constitutive androstane receptor (CAR) and FXR seem to be responsible for the induction of hBD-1 by bilirubin.
Conclusion: hBD-1 is prominently expressed in hepatocytes. It is induced during cholestasis through bilirubin and BAs, mediated by CAR and especially FXR. Reduction by TXN activates hBD-1 to a potential key player in innate antimicrobial defense of the liver.
Keywords: antimicrobial peptides; bilirubin; cholestasis; hepatocytes; human β-defensin-1.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

