Container-based bioinformatics with Pachyderm
- PMID: 30101309
- PMCID: PMC6394392
- DOI: 10.1093/bioinformatics/bty699
Container-based bioinformatics with Pachyderm
Abstract
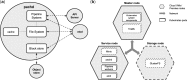
Motivation: Computational biologists face many challenges related to data size, and they need to manage complicated analyses often including multiple stages and multiple tools, all of which must be deployed to modern infrastructures. To address these challenges and maintain reproducibility of results, researchers need (i) a reliable way to run processing stages in any computational environment, (ii) a well-defined way to orchestrate those processing stages and (iii) a data management layer that tracks data as it moves through the processing pipeline.
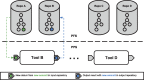
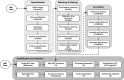
Results: Pachyderm is an open-source workflow system and data management framework that fulfils these needs by creating a data pipelining and data versioning layer on top of projects from the container ecosystem, having Kubernetes as the backbone for container orchestration. We adapted Pachyderm and demonstrated its attractive properties in bioinformatics. A Helm Chart was created so that researchers can use Pachyderm in multiple scenarios. The Pachyderm File System was extended to support block storage. A wrapper for initiating Pachyderm on cloud-agnostic virtual infrastructures was created. The benefits of Pachyderm are illustrated via a large metabolomics workflow, demonstrating that Pachyderm enables efficient and sustainable data science workflows while maintaining reproducibility and scalability.
Availability and implementation: Pachyderm is available from https://github.com/pachyderm/pachyderm. The Pachyderm Helm Chart is available from https://github.com/kubernetes/charts/tree/master/stable/pachyderm. Pachyderm is available out-of-the-box from the PhenoMeNal VRE (https://github.com/phnmnl/KubeNow-plugin) and general Kubernetes environments instantiated via KubeNow. The code of the workflow used for the analysis is available on GitHub (https://github.com/pharmbio/LC-MS-Pachyderm).
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2018. Published by Oxford University Press.
Figures

References
-
- Barba L.A. (2016) The hard road to reproducibility. Science, 354, 142.. - PubMed
-
- Begley C.G., Ioannidis J.P. (2015) Reproducibility in science: improving the standard for basic and preclinical research. Circ. Res., 116, 116–126. - PubMed
-
- Burns B., Oppenheimer D. (2016) Design patterns for container-based distributed systems. In: HotCloud’16 Proceedings of the 8th USENIX Conference on Hot Topics in Cloud Computing. pp. 108–113. USENIX Association, Berkeley, CA, USA.
-
- Capuccini M. et al. (2018) KubeNow: a cloud agnostic platform for microservice-oriented applications In: Leahy F., Franco J. (eds), 2017 Imperial College Computing Student Workshop (ICCSW 2017), Volume 60 of OpenAccess Series in Informatics (OASIcs). Schloss Dagstuhl–Leibniz-Zentrum fuer Informatik, Germany, pp. 9:1–9:2.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

