RNA Structure and Cellular Applications of Fluorescent Light-Up Aptamers
- PMID: 30102012
- PMCID: PMC6391945
- DOI: 10.1002/anie.201806482
RNA Structure and Cellular Applications of Fluorescent Light-Up Aptamers
Abstract
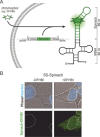
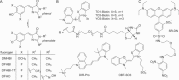
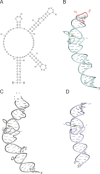
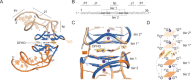
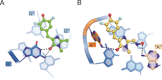
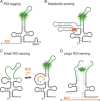
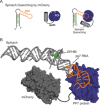
The cellular functions of RNA are not limited to their role as blueprints for protein synthesis. In particular, noncoding RNA, such as, snRNAs, lncRNAs, miRNAs, play important roles. With increasing numbers of RNAs being identified, it is well known that the transcriptome outnumbers the proteome by far. This emphasizes the great importance of functional RNA characterization and the need to further develop tools for these investigations, many of which are still in their infancy. Fluorescent light-up aptamers (FLAPs) are RNA sequences that can bind nontoxic, cell-permeable small-molecule fluorogens and enhance their fluorescence over many orders of magnitude upon binding. FLAPs can be encoded on the DNA level using standard molecular biology tools and are subsequently transcribed into RNA by the cellular machinery, so that they can be used as fluorescent RNA tags (FLAP-tags). In this Minireview, we give a brief overview of the fluorogens that have been developed and their binding RNA aptamers, with a special focus on published crystal structures. A summary of current and future cellular FLAP applications with an emphasis on the study of RNA-RNA and RNA-protein interactions using split-FLAP and Förster resonance energy transfer (FRET) systems is given.
Keywords: RNA imaging; RNA structures; RNA-protein interactions; fluorescent light-up aptamers (FLAPs); fluorogenic probes.
© 2018 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
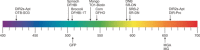



References
-
- König J., Zarnack K., Luscombe N. M., Ule J., Nat. Rev. Genet. 2012, 13, 77–83. - PubMed
-
- Mercer T. R., Dinger M. E., Mattick J. S., Nat. Rev. Genet. 2009, 10, 155–159. - PubMed
-
- McHugh C. A., Russell P., Guttman M., Chen M., Manley J., Licatalosi D., Darnell R., Wang Z., Gerstein M., Snyder M., et al., Genome Biol. 2014, 15, 1–10.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

