Impulse model-based differential expression analysis of time course sequencing data
- PMID: 30102402
- PMCID: PMC6237758
- DOI: 10.1093/nar/gky675
Impulse model-based differential expression analysis of time course sequencing data
Abstract
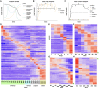
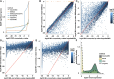
Temporal changes to the concentration of molecular species such as mRNA, which take place in response to various environmental cues, can often be modeled as simple continuous functions such as a single pulse (impulse) model. The simplicity of such functional representations can provide an improved performance on fundamental tasks such as noise reduction, imputation and differential expression analysis. However, temporal gene expression profiles are often studied with models that treat time as a categorical variable, neglecting the dependence between time points. Here, we present ImpulseDE2, a framework for differential expression analysis that combines the power of the impulse model as a continuous representation of temporal responses along with a noise model tailored specifically to sequencing data. We compare the simple categorical models to ImpulseDE2 and to other continuous models based on natural cubic splines and demonstrate the utility of the continuous approach for studying differential expression in time course sequencing experiments. A unique feature of ImpulseDE2 is the ability to distinguish permanently from transiently up- or down-regulated genes. Using an in vitro differentiation dataset, we demonstrate that this gene classification scheme can be used to highlight distinct transcriptional programs that are associated with different phases of the differentiation process.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

