RbpA relaxes promoter selectivity of M. tuberculosis RNA polymerase
- PMID: 30102406
- PMCID: PMC6212719
- DOI: 10.1093/nar/gky714
RbpA relaxes promoter selectivity of M. tuberculosis RNA polymerase
Abstract
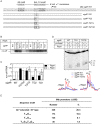
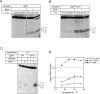
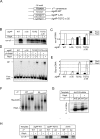
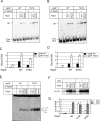
The transcriptional activator RbpA associates with Mycobacterium tuberculosis RNA polymerase (MtbRNAP) during transcription initiation, and stimulates formation of the MtbRNAP-promoter open complex (RPo). Here, we explored the influence of promoter motifs on RbpA-mediated activation of MtbRNAP containing the stress-response σB subunit. We show that both the 'extended -10' promoter motif (T-17G-16T-15G-14) and RbpA stabilized RPo and allowed promoter opening at suboptimal temperatures. Furthermore, in the presence of the T-17G-16T-15G-14 motif, RbpA was dispensable for RNA synthesis initiation, while exerting a stabilization effect on RPo. On the other hand, RbpA compensated for the lack of sequence-specific interactions of domains 3 and 4 of σB with the extended -10 and the -35 motifs, respectively. Mutations of the positively charged residues K73, K74 and R79 in RbpA basic linker (BL) had little effect on RPo formation, but affected MtbRNAP capacity for de novo transcription initiation. We propose that RbpA stimulates transcription by strengthening the non-specific interaction of the σ subunit with promoter DNA upstream of the -10 element, and by indirectly optimizing MtbRNAP interaction with initiation substrates. Consequently, RbpA renders MtbRNAP promiscuous in promoter selection, thus compensating for the weak conservation of the -35 motif in mycobacteria.
Figures



References
-
- Feklístov A., Sharon B.D., Darst S.A., Gross C.A.. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014; 68:357–376. - PubMed
-
- Borukhov S., Nudler E.. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008; 16:126–134. - PubMed
-
- Rodrigue S., Provvedi R., Jacques P., Gaudreau L., Manganelli R.. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2006; 30:926–941. - PubMed
-
- Gruber T.M., Gross C.A.. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003; 57:441–466. - PubMed
-
- Graves M.C., Rabinowitz J.C.. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for “extended” promoter elements in gram-positive organisms. J. Biol. Chem. 1986; 261:11409–11415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

