Rapid Ion Mobility Separations of Bile Acid Isomers Using Cyclodextrin Adducts and Structures for Lossless Ion Manipulations
- PMID: 30102518
- PMCID: PMC6191033
- DOI: 10.1021/acs.analchem.8b02990
Rapid Ion Mobility Separations of Bile Acid Isomers Using Cyclodextrin Adducts and Structures for Lossless Ion Manipulations
Abstract
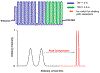
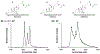
Bile acids (BAs) constitute an important class of steroid metabolites often displaying changes associated with disease states and other health conditions. Current analyses for these structurally similar compounds are limited by a lack of sensitivity and long separation times with often poor isomeric resolution. To overcome these challenges and provide rapid analyses for the BA isomers, we utilized cyclodextrin adducts in conjunction with novel ion mobility (IM) separation capabilities provided by structures for lossless ion manipulations (SLIM). Cyclodextrin was found to interact with both the tauro- and glyco-conjugated BA isomers studied, forming rigid noncovalent host-guest inclusion complexes. Without the use of cyclodextrin adducts, the BA isomers were found to be nearly identical in their respective mobilities and thus unable to be baseline resolved. Each separation of the cyclodextrin-bile acid host-guest inclusion complex was performed in less than 1 s, providing a much more rapid alternative to current liquid chromatography-based separations. SLIM provided capabilities for the accumulation of larger ion populations and IM peak compression that resulted in much higher resolution separations and increased signal intensities for the BA isomers studied.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Chen M; Gratzel M; Thomas JK J. Am. Chem. Soc 1975, 97 (8), 2052–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

