Scaling up data curation using deep learning: An application to literature triage in genomic variation resources
- PMID: 30102703
- PMCID: PMC6107285
- DOI: 10.1371/journal.pcbi.1006390
Scaling up data curation using deep learning: An application to literature triage in genomic variation resources
Abstract
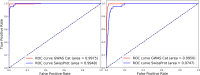
Manually curating biomedical knowledge from publications is necessary to build a knowledge based service that provides highly precise and organized information to users. The process of retrieving relevant publications for curation, which is also known as document triage, is usually carried out by querying and reading articles in PubMed. However, this query-based method often obtains unsatisfactory precision and recall on the retrieved results, and it is difficult to manually generate optimal queries. To address this, we propose a machine-learning assisted triage method. We collect previously curated publications from two databases UniProtKB/Swiss-Prot and the NHGRI-EBI GWAS Catalog, and used them as a gold-standard dataset for training deep learning models based on convolutional neural networks. We then use the trained models to classify and rank new publications for curation. For evaluation, we apply our method to the real-world manual curation process of UniProtKB/Swiss-Prot and the GWAS Catalog. We demonstrate that our machine-assisted triage method outperforms the current query-based triage methods, improves efficiency, and enriches curated content. Our method achieves a precision 1.81 and 2.99 times higher than that obtained by the current query-based triage methods of UniProtKB/Swiss-Prot and the GWAS Catalog, respectively, without compromising recall. In fact, our method retrieves many additional relevant publications that the query-based method of UniProtKB/Swiss-Prot could not find. As these results show, our machine learning-based method can make the triage process more efficient and is being implemented in production so that human curators can focus on more challenging tasks to improve the quality of knowledge bases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
On expert curation and scalability: UniProtKB/Swiss-Prot as a case study.Bioinformatics. 2017 Nov 1;33(21):3454-3460. doi: 10.1093/bioinformatics/btx439. Bioinformatics. 2017. PMID: 29036270 Free PMC article.
-
An enhanced workflow for variant interpretation in UniProtKB/Swiss-Prot improves consistency and reuse in ClinVar.Database (Oxford). 2019 Jan 1;2019:baz040. doi: 10.1093/database/baz040. Database (Oxford). 2019. PMID: 30937429 Free PMC article.
-
Using distant supervised learning to identify protein subcellular localizations from full-text scientific articles.J Biomed Inform. 2015 Oct;57:134-44. doi: 10.1016/j.jbi.2015.07.013. Epub 2015 Jul 26. J Biomed Inform. 2015. PMID: 26220461
-
Genetic variations and diseases in UniProtKB/Swiss-Prot: the ins and outs of expert manual curation.Hum Mutat. 2014 Aug;35(8):927-35. doi: 10.1002/humu.22594. Epub 2014 Jun 24. Hum Mutat. 2014. PMID: 24848695 Free PMC article. Review.
-
Challenges in the annotation of pseudoenzymes in databases: the UniProtKB approach.FEBS J. 2020 Oct;287(19):4114-4127. doi: 10.1111/febs.15100. Epub 2019 Nov 3. FEBS J. 2020. PMID: 31618524 Free PMC article. Review.
Cited by
-
Recent advances of automated methods for searching and extracting genomic variant information from biomedical literature.Brief Bioinform. 2021 May 20;22(3):bbaa142. doi: 10.1093/bib/bbaa142. Brief Bioinform. 2021. PMID: 32770181 Free PMC article. Review.
-
LitSuggest: a web-based system for literature recommendation and curation using machine learning.Nucleic Acids Res. 2021 Jul 2;49(W1):W352-W358. doi: 10.1093/nar/gkab326. Nucleic Acids Res. 2021. PMID: 33950204 Free PMC article.
-
BioConceptVec: Creating and evaluating literature-based biomedical concept embeddings on a large scale.PLoS Comput Biol. 2020 Apr 23;16(4):e1007617. doi: 10.1371/journal.pcbi.1007617. eCollection 2020 Apr. PLoS Comput Biol. 2020. PMID: 32324731 Free PMC article.
-
Using deep learning to identify translational research in genomic medicine beyond bench to bedside.Database (Oxford). 2019 Jan 1;2019:baz010. doi: 10.1093/database/baz010. Database (Oxford). 2019. PMID: 30753477 Free PMC article.
-
UPCLASS: a deep learning-based classifier for UniProtKB entry publications.Database (Oxford). 2020 Jan 1;2020:baaa026. doi: 10.1093/database/baaa026. Database (Oxford). 2020. PMID: 32367111 Free PMC article.
References
-
- Famiglietti ML, Estreicher A, Gos A, Bolleman J, Gehant S, Breuza L, et al. Genetic variations and diseases in UniProtKB/Swiss-Prot: the ins and outs of expert manual curation. Hum Mutat. 2014;35(8):927–35. Epub 2014/05/23. 10.1002/humu.22594 ; PubMed Central PMCID: PMCPMC4107114. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials