Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains
- PMID: 30102851
- PMCID: PMC7168555
- DOI: 10.1111/tbed.12991
Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains
Abstract
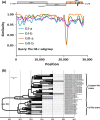
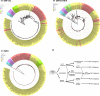
Porcine epidemic diarrhea virus (PEDV), which re-emerged in China in October 2010, has spread rapidly worldwide. Detailed analyses of the complete genomes of different PEDV strains are essential to understand the relationships among re-emerging and historic strains worldwide. Here, we analysed the complete genomes of 409 strains from different countries, which were classified into five subgroup strains (i.e., GI-a, GI-b, GII-a, GII-b, and GII-c). Phylogenetic study of different genes in the PEDV strains revealed that the newly discovered subgroup GII-c exhibited inconsistent topologies between the spike gene and other genes. Furthermore, recombination analysis indicated that GII-c viruses evolved from a recombinant virus that acquired the 5' part of the spike gene from the GI-a subgroup and the remaining genomic regions from the GII-a subgroup. Molecular clock analysis showed that divergence of the GII-c subgroup spike gene occurred in April 2010, suggesting that the subgroup originated from recombination events before the PEDV re-emergence outbreaks. Interestingly, Ascaris suum, a large roundworm occurring in pigs, was found to be an unusual PEDV host, providing potential support for cross-host transmission. This study has significant implications for understanding ongoing global PEDV outbreaks and will guide future efforts to develop effective preventative measures against PEDV.
Keywords: genotyping; molecular epidemiology; porcine epidemic diarrhoea virus; recombination; spike gene.
© 2018 Blackwell Verlag GmbH.
Conflict of interest statement
None.
Figures


References
-
- Chang, S. H. , Bae, J. L. , Kang, T. J. , Kim, J. , Chung, G. H. , Lim, C. W. , … Jang, Y. S. (2002). Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Molecules and cells, 14, 295–299. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- 31672569, 31672566/National Natural Science Foundation of China
- 2016YFD0500103/National Key R&D Program of China
- 2017CFA059/Natural Science Foundation of Hubei Province
- 2017ABA138/Special Project for Technology Innovation of Hubei Province
- 2662016PY070/Fundamental Research Funds for the Central Universities
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

