Postsynaptic Proteome of Non-Demented Individuals with Alzheimer's Disease Neuropathology
- PMID: 30103319
- PMCID: PMC6130411
- DOI: 10.3233/JAD-180179
Postsynaptic Proteome of Non-Demented Individuals with Alzheimer's Disease Neuropathology
Abstract
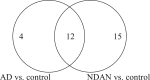
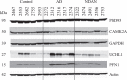
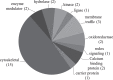
Some individuals, here referred to as Non-Demented with Alzheimer's Neuropathology (NDAN), retain their cognitive function despite the presence of amyloid plaques and tau tangles typical of symptomatic Alzheimer's disease (AD). In NDAN, unlike AD, toxic amyloid-β oligomers do not localize to the postsynaptic densities (PSDs). Synaptic resistance to amyloid-β in NDAN may thus enable these individuals to remain cognitively intact despite the AD-like pathology. The mechanism(s) responsible for this resistance remains unresolved and understanding such protective biological processes could reveal novel targets for the development of effective treatments for AD. The present study uses a proteomic approach to compare the hippocampal postsynaptic densities of NDAN, AD, and healthy age-matched persons to identify protein signatures characteristic for these groups. Subcellular fractionation followed by 2D gel electrophoresis and mass spectrometry were used to analyze the PSDs. We describe fifteen proteins which comprise the unique proteomic signature of NDAN PSDs, thus setting them apart from control subjects and AD patients.
Keywords: Alzheimer’s disease; non-demented with AD-like pathology; postsynaptic density proteome; synapse.
Figures

References
-
- Alzheimer’s Association (2017) 2017 Alzheimer’s disease facts and figures, Alzheimers Dement 13, 325–373.
-
- Assessing Risk for Alzheimer’s Disease, https://www.nia.nih.gov/health/assessing-risk-alzheimers-disease, Last updated May 19, 2017, Accessed on July 06, 2017.
-
- Matrone C, Djelloul M, Taglialatela G, Perrone L (2015) Inflammatory risk factors and pathologies promoting Alzheimer’s disease progression: Is RAGE the key?, Histol Histopathol 30, 125–139. - PubMed
-
- Aluise CD, Robinson RA, Cai J, Pierce WM, Markesbery WR, Butterfield DA (2011) Redox proteomics analysis of brains from subjects with amnestic mild cognitive impairment compared to brains from subjects with preclinical Alzheimer’s disease: Insights into memory loss in MCI, J Alzheimers Dis 23, 257–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

