The tick endosymbiont Candidatus Midichloria mitochondrii and selenoproteins are essential for the growth of Rickettsia parkeri in the Gulf Coast tick vector
- PMID: 30103809
- PMCID: PMC6090677
- DOI: 10.1186/s40168-018-0524-2
The tick endosymbiont Candidatus Midichloria mitochondrii and selenoproteins are essential for the growth of Rickettsia parkeri in the Gulf Coast tick vector
Abstract
Background: Pathogen colonization inside tick tissues is a significant aspect of the overall competence of a vector. Amblyomma maculatum is a competent vector of the spotted fever group rickettsiae, Rickettsia parkeri. When R. parkeri colonizes its tick host, it has the opportunity to dynamically interact with not just its host but with the endosymbionts living within it, and this enables it to modulate the tick's defenses by regulating tick gene expression. The microbiome in A. maculatum is dominated by two endosymbiont microbes: a Francisella-like endosymbiont (FLE) and Candidatus Midichloria mitochondrii (CMM). A range of selenium-containing proteins (selenoproteins) in A. maculatum ticks protects them from oxidative stress during blood feeding and pathogen infections. Here, we investigated rickettsial multiplication in the presence of tick endosymbionts and characterized the functional significance of selenoproteins during R. parkeri replication in the tick.
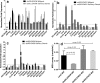
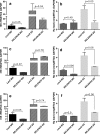
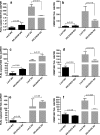
Results: FLE and CMM were quantified throughout the tick life stages by quantitative PCR in R. parkeri-infected and uninfected ticks. R. parkeri infection was found to decrease the FLE numbers but CMM thrived across the tick life cycle. Our qRT-PCR analysis indicated that the transcripts of genes with functions related to redox (selenogenes) were upregulated in ticks infected with R. parkeri. Three differentially expressed proteins, selenoprotein M, selenoprotein O, and selenoprotein S were silenced to examine their functional significance during rickettsial replication within the tick tissues. Gene silencing of the target genes was found to impair R. parkeri colonization in the tick vector. Knockdown of the selenogenes triggered a compensatory response from other selenogenes, as observed by changes in gene expression, but oxidative stress levels and endoplasmic reticulum stress inside the ticks were also found to have heightened.
Conclusions: This study illustrates the potential of this new research model for augmenting our understanding of the pathogen interactions occurring within tick hosts and the important roles that symbionts and various tick factors play in regulating pathogen growth.
Keywords: Colonization; Endosymbionts; Pathogen; Rickettsia parkeri; Selenogenes; Ticks.
Conflict of interest statement
Use of animals for tick blood-feeding was approved by the IACUC of the University of Southern Mississippi.
The authors declare that they have no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
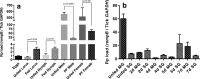

Similar articles
-
Hematophagy and tick-borne Rickettsial pathogen shape the microbial community structure and predicted functions within the tick vector, Amblyomma maculatum.Front Cell Infect Microbiol. 2022 Nov 21;12:1037387. doi: 10.3389/fcimb.2022.1037387. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36478675 Free PMC article.
-
Rickettsia parkeri colonization in Amblyomma maculatum: the role of superoxide dismutases.Parasit Vectors. 2016 May 20;9(1):291. doi: 10.1186/s13071-016-1579-1. Parasit Vectors. 2016. PMID: 27206371 Free PMC article.
-
Amblyomma maculatum SECIS binding protein 2 and putative selenoprotein P are indispensable for pathogen replication and tick fecundity.Insect Biochem Mol Biol. 2017 Sep;88:37-47. doi: 10.1016/j.ibmb.2017.07.006. Epub 2017 Jul 21. Insect Biochem Mol Biol. 2017. PMID: 28739494 Free PMC article.
-
Recent advances in understanding tick and rickettsiae interactions.Parasite Immunol. 2021 May;43(5):e12830. doi: 10.1111/pim.12830. Epub 2021 Apr 15. Parasite Immunol. 2021. PMID: 33713348 Free PMC article. Review.
-
Impact of endosymbionts on tick physiology and fitness.Parasitology. 2023 Sep;150(10):859-865. doi: 10.1017/S0031182023000793. Epub 2023 Aug 24. Parasitology. 2023. PMID: 37722758 Free PMC article. Review.
Cited by
-
Ambivalent Roles of Oxidative Stress in Triangular Relationships among Arthropod Vectors, Pathogens and Hosts.Antioxidants (Basel). 2022 Jun 25;11(7):1254. doi: 10.3390/antiox11071254. Antioxidants (Basel). 2022. PMID: 35883744 Free PMC article. Review.
-
Multi-country investigation of the diversity and associated microorganisms isolated from tick species from domestic animals, wildlife and vegetation in selected african countries.Exp Appl Acarol. 2021 Mar;83(3):427-448. doi: 10.1007/s10493-021-00598-3. Epub 2021 Mar 1. Exp Appl Acarol. 2021. PMID: 33646482 Free PMC article.
-
The Symbiotic Continuum Within Ticks: Opportunities for Disease Control.Front Microbiol. 2022 Mar 17;13:854803. doi: 10.3389/fmicb.2022.854803. eCollection 2022. Front Microbiol. 2022. PMID: 35369485 Free PMC article. Review.
-
Rickettsia parkeri infection modulates the sialome and ovariome of the Gulf coast tick, Amblyomma maculatum.Front Microbiol. 2022 Nov 10;13:1023980. doi: 10.3389/fmicb.2022.1023980. eCollection 2022. Front Microbiol. 2022. PMID: 36439862 Free PMC article.
-
Hematophagy and tick-borne Rickettsial pathogen shape the microbial community structure and predicted functions within the tick vector, Amblyomma maculatum.Front Cell Infect Microbiol. 2022 Nov 21;12:1037387. doi: 10.3389/fcimb.2022.1037387. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36478675 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

