The newly-arisen Devil facial tumour disease 2 (DFT2) reveals a mechanism for the emergence of a contagious cancer
- PMID: 30103855
- PMCID: PMC6092122
- DOI: 10.7554/eLife.35314
The newly-arisen Devil facial tumour disease 2 (DFT2) reveals a mechanism for the emergence of a contagious cancer
Abstract
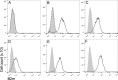
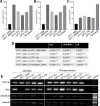
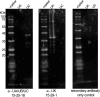
Devil Facial Tumour 2 (DFT2) is a recently discovered contagious cancer circulating in the Tasmanian devil (Sarcophilus harrisii), a species which already harbours a more widespread contagious cancer, Devil Facial Tumour 1 (DFT1). Here we show that in contrast to DFT1, DFT2 cells express major histocompatibility complex (MHC) class I molecules, demonstrating that loss of MHC is not necessary for the emergence of a contagious cancer. However, the most highly expressed MHC class I alleles in DFT2 cells are common among host devils or non-polymorphic, reducing immunogenicity in a population sharing these alleles. In parallel, MHC class I loss is emerging in vivo, thus DFT2 may be mimicking the evolutionary trajectory of DFT1. Based on these results we propose that contagious cancers may exploit partial histocompatibility between the tumour and host, but that loss of allogeneic antigens could facilitate widespread transmission of DFT2.
Keywords: MHC; Sarcophilus harrisii; cancer biology; contagious cancer; immune escape; immunology; inflammation.
© 2018, Caldwell et al.
Conflict of interest statement
AC, RC, CT, MS, YK, RO, MT, EM, KS, GT, JK, TE, GW, HS No competing interests declared
Figures






Comment in
-
A 'devil' of a problem.Elife. 2018 Aug 14;7:e39976. doi: 10.7554/eLife.39976. Elife. 2018. PMID: 30103857 Free PMC article.
References
-
- Almand B, Clark JI, Nikitina E, van Beynen J, English NR, Knight SC, Carbone DP, Gabrilovich DI. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. The Journal of Immunology. 2001;166:678–689. doi: 10.4049/jimmunol.166.1.678. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

