In Vivo Virulence Characterization of Pregnancy-Associated Listeria monocytogenes Infections
- PMID: 30104213
- PMCID: PMC6204711
- DOI: 10.1128/IAI.00397-18
In Vivo Virulence Characterization of Pregnancy-Associated Listeria monocytogenes Infections
Abstract
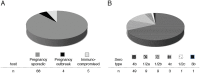
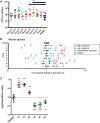
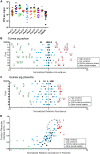
Listeria monocytogenes is a foodborne pathogen that infects the placenta and can cause pregnancy complications. Listeriosis usually occurs as a sporadic infection, but large outbreaks are also reported. Virulence from clinical isolates is rarely analyzed due to the large number of animals required, but this knowledge could help guide the response to an outbreak. We implemented a DNA barcode system using signature tags that allowed us to efficiently assay variations in virulence across a large number of isolates. We tested 77 signature-tagged clones of clinical L. monocytogenes strains from 72 infected human placentas and 5 immunocompromised patients, all of which were isolated since 2000. These strains were tested for virulence in a modified competition assay in comparison to that of the laboratory strain 10403S. We used two in vivo models of listeriosis: the nonpregnant mouse and the pregnant guinea pig. Strains that were frequently found at a high abundance within infected organs were considered hypervirulent, while strains frequently found at a low abundance were considered hypovirulent. Virulence split relatively evenly among hypovirulent strains, hypervirulent strains, and strains as virulent as 10403S. The laboratory strain was found to have an intermediate virulence phenotype, supporting its suitability for use in pathogenesis studies. Further, we found that splenic virulence and placental virulence are closely linked in both the guinea pig and mouse models. This suggests that outbreak and sporadic pregnancy-associated L. monocytogenes strains are not generally more virulent than lab reference strains. However, some strains did show consistent and reproducible virulence differences, suggesting that their further study may reveal deeper insights into the biological underpinnings of listeriosis.
Keywords: DNA barcode; Listeria; clinical isolate; epidemiology; placental infection; placental pathogen; signature tag; virulence.
Copyright © 2018 Morrison et al.
Figures
Similar articles
-
A processing plant persistent strain of Listeria monocytogenes crosses the fetoplacental barrier in a pregnant guinea pig model.J Food Prot. 2008 May;71(5):1028-34. doi: 10.4315/0362-028x-71.5.1028. J Food Prot. 2008. PMID: 18522041
-
Pregnancy - associated human listeriosis: Virulence and genotypic analysis of Listeria monocytogenes from clinical samples.J Microbiol. 2015 Sep;53(9):653-60. doi: 10.1007/s12275-015-5243-9. Epub 2015 Aug 1. J Microbiol. 2015. PMID: 26231373
-
InlP, a New Virulence Factor with Strong Placental Tropism.Infect Immun. 2016 Nov 18;84(12):3584-3596. doi: 10.1128/IAI.00625-16. Print 2016 Dec. Infect Immun. 2016. PMID: 27736782 Free PMC article.
-
Virulence characterization of Listeria monocytogenes.Methods Mol Biol. 2014;1157:157-65. doi: 10.1007/978-1-4939-0703-8_13. Methods Mol Biol. 2014. PMID: 24792556 Review.
-
Maternal-neonatal listeriosis.Virulence. 2020 Dec;11(1):391-397. doi: 10.1080/21505594.2020.1759287. Virulence. 2020. PMID: 32363991 Free PMC article. Review.
Cited by
-
Infections at the maternal-fetal interface: an overview of pathogenesis and defence.Nat Rev Microbiol. 2022 Feb;20(2):67-82. doi: 10.1038/s41579-021-00610-y. Epub 2021 Aug 25. Nat Rev Microbiol. 2022. PMID: 34433930 Free PMC article. Review.
-
Enrichment of Neutrophils and Monocytes From the Liver Following Either Oral or Intravenous Listeria monocytogenes Infection.Curr Protoc Immunol. 2020 Sep;130(1):e102. doi: 10.1002/cpim.102. Curr Protoc Immunol. 2020. PMID: 32710703 Free PMC article.
-
Listeria monocytogenes infection in pregnant macaques alters the maternal gut microbiome†.Biol Reprod. 2023 Nov 15;109(5):618-634. doi: 10.1093/biolre/ioad104. Biol Reprod. 2023. PMID: 37665249 Free PMC article.
-
TMT-Based Quantitative Proteomic Analysis of Intestinal Organoids Infected by Listeria monocytogenes Strains with Different Virulence.Int J Mol Sci. 2022 Jun 2;23(11):6231. doi: 10.3390/ijms23116231. Int J Mol Sci. 2022. PMID: 35682909 Free PMC article.
-
Sow reproductive disorders: a key issue affecting the pig industry.Front Vet Sci. 2025 Mar 5;12:1535719. doi: 10.3389/fvets.2025.1535719. eCollection 2025. Front Vet Sci. 2025. PMID: 40110434 Free PMC article. Review.
References
-
- Lorber B. 1997. Listeriosis. Clin Infect Dis 24:1–9. - PubMed
-
- McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O'Connor KA, Cosgrove S, Gossack JP, Parachini SS, Jain NS, Ettestad P, Ibraheem M, Cantu V, Joshi M, DuVernoy T, Fogg NW Jr, Gorny JR, Mogen KM, Spires C, Teitell P, Joseph LA, Tarr CL, Imanishi M, Neil KP, Tauxe RV, Mahon BE. 2013. Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med 369:944–953. doi:10.1056/NEJMoa1215837. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

