RNF169 limits 53BP1 deposition at DSBs to stimulate single-strand annealing repair
- PMID: 30104380
- PMCID: PMC6126738
- DOI: 10.1073/pnas.1804823115
RNF169 limits 53BP1 deposition at DSBs to stimulate single-strand annealing repair
Abstract
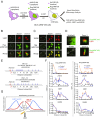
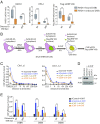
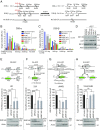
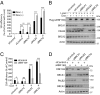
Unrestrained 53BP1 activity at DNA double-strand breaks (DSBs) hampers DNA end resection and upsets DSB repair pathway choice. RNF169 acts as a molecular rheostat to limit 53BP1 deposition at DSBs, but how this fine balance translates to DSB repair control remains undefined. In striking contrast to 53BP1, ChIP analyses of AsiSI-induced DSBs unveiled that RNF169 exhibits robust accumulation at DNA end-proximal regions and preferentially targets resected, RPA-bound DSBs. Accordingly, we found that RNF169 promotes CtIP-dependent DSB resection and favors homology-mediated DSB repair, and further showed that RNF169 dose-dependently stimulates single-strand annealing repair, in part, by alleviating the 53BP1-imposed barrier to DSB end resection. Our results highlight the interplay of RNF169 with 53BP1 in fine-tuning choice of DSB repair pathways.
Keywords: 53BP1; DNA damage; DNA double-strand breaks; RNF169; single-strand annealing repair.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mani RS, Chinnaiyan AM. Triggers for genomic rearrangements: Insights into genomic, cellular and environmental influences. Nat Rev Genet. 2010;11:819–829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

