Evaluating the Functional Pore Size of Chloroplast TOC and TIC Protein Translocons: Import of Folded Proteins
- PMID: 30104404
- PMCID: PMC6181021
- DOI: 10.1105/tpc.18.00427
Evaluating the Functional Pore Size of Chloroplast TOC and TIC Protein Translocons: Import of Folded Proteins
Abstract
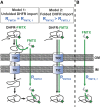
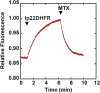
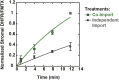
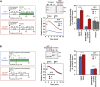
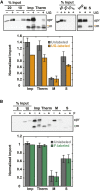
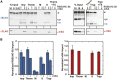
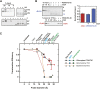
The degree of residual structure retained by proteins while passing through biological membranes is a fundamental mechanistic question of protein translocation. Proteins are generally thought to be unfolded while transported through canonical proteinaceous translocons, including the translocons of the outer and inner chloroplast envelope membranes (TOC and TIC). Here, we readdressed the issue and found that the TOC/TIC translocons accommodated the tightly folded dihydrofolate reductase (DHFR) protein in complex with its stabilizing ligand, methotrexate (MTX). We employed a fluorescein-conjugated methotrexate (FMTX), which has slow membrane transport rates relative to unconjugated MTX, to show that the rate of ligand accumulation inside chloroplasts is faster when bound to DHFR that is actively being imported. Stromal accumulation of FMTX is ATP dependent when DHFR is actively being imported but is otherwise ATP independent, again indicating DHFR/FMTX complex import. Furthermore, the TOC/TIC pore size was probed with fixed-diameter particles and found to be greater than 25.6 Å, large enough to support folded DHFR import and also larger than mitochondrial and bacterial protein translocons that have a requirement for protein unfolding. This unique pore size and the ability to import folded proteins have critical implications regarding the structure and mechanism of the TOC/TIC translocons.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures


References
-
- America T., Hageman J., Guéra A., Rook F., Archer K., Keegstra K., Weisbeek P. (1994). Methotrexate does not block import of a DHFR fusion protein into chloroplasts. Plant Mol. Biol. 24: 283–294. - PubMed
-
- Assaraf Y.G., Seamer L.C., Schimke R.T. (1989). Characterization by flow cytometry of fluorescein-methotrexate transport in Chinese hamster ovary cells. Cytometry 10: 50–55. - PubMed
-
- Baud C., Guérin J., Petit E., Lesne E., Dupré E., Locht C., Jacob-Dubuisson F. (2014). Translocation path of a substrate protein through its Omp85 transporter. Nat. Commun. 5: 5271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

