Detecting Rare Mutations with Heterogeneous Effects Using a Family-Based Genetic Random Field Method
- PMID: 30104420
- PMCID: PMC6216585
- DOI: 10.1534/genetics.118.301266
Detecting Rare Mutations with Heterogeneous Effects Using a Family-Based Genetic Random Field Method
Abstract
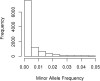
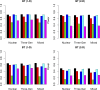
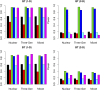
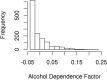
The genetic etiology of many complex diseases is highly heterogeneous. A complex disease can be caused by multiple mutations within the same gene or mutations in multiple genes at various genomic loci. Although these disease-susceptibility mutations can be collectively common in the population, they are often individually rare or even private to certain families. Family-based studies are powerful for detecting rare variants enriched in families, which is an important feature for sequencing studies due to the heterogeneous nature of rare variants. In addition, family designs can provide robust protection against population stratification. Nevertheless, statistical methods for analyzing family-based sequencing data are underdeveloped, especially those accounting for heterogeneous etiology of complex diseases. In this article, we introduce a random field framework for detecting gene-phenotype associations in family-based sequencing studies, referred to as family-based genetic random field (FGRF). Similar to existing family-based association tests, FGRF could utilize within-family and between-family information separately or jointly to test an association. We demonstrate that FGRF has comparable statistical power with existing methods when there is no genetic heterogeneity, but can improve statistical power when there is genetic heterogeneity across families. The proposed method also shares the same advantages with the conventional family-based association tests (e.g., being robust to population stratification). Finally, we applied the proposed method to a sequencing data from the Minnesota Twin Family Study, and revealed several genes, including SAMD14, potentially associated with alcohol dependence.
Keywords: alcohol dependence; family-based association study; genetic heterogeneity; population stratification; rare variants.
Copyright © 2018 by the Genetics Society of America.
Figures


References
-
- Adler R. J., Taylor J. E., 2007. Random Field and Geometry. Springer, New York.
-
- Argani P., Iacobuzio-Donahue C., Ryu B., Rosty C., Goggins M., et al. , 2001. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE). Clin. Cancer Res. 7: 3862–3868. - PubMed
-
- Berg K. A., Astemborski J. A., Boughman J. A., Ferencz C., 1989. Congenital cardiovascular malformations in twins and triplets from a population-based study. Am. J. Dis. Child. 143: 1461–1463. - PubMed
-
- Boos D. D., 1992. On generalized score tests. Am. Stat. 46: 327–333.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

