Visual and modular detection of pathogen nucleic acids with enzyme-DNA molecular complexes
- PMID: 30104566
- PMCID: PMC6089962
- DOI: 10.1038/s41467-018-05733-0
Visual and modular detection of pathogen nucleic acids with enzyme-DNA molecular complexes
Abstract
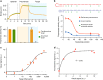
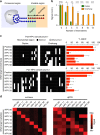
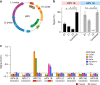
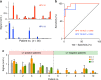
Rapid, visual detection of pathogen nucleic acids has broad applications in infection management. Here we present a modular detection platform, termed enzyme-assisted nanocomplexes for visual identification of nucleic acids (enVision). The system consists of an integrated circuit of enzyme-DNA nanostructures, which function as independent recognition and signaling elements, for direct and versatile detection of pathogen nucleic acids from infected cells. The built-in enzymatic cascades produce a rapid color readout for the naked eye; the assay is thus fast (<2 h), sensitive (<10 amol), and readily quantified with smartphones. When implemented on a configurable microfluidic platform, the technology demonstrates superior programmability to perform versatile computations, for detecting diverse pathogen targets and their virus-host genome integration loci. We further design the enVision platform for molecular-typing of infections in patient endocervical samples. The technology not only improves the clinical inter-subtype differentiation, but also expands the intra-subtype coverage to identify previously undetectable infections.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

