Determining Relative Dynamic Stability of Cell States Using Boolean Network Model
- PMID: 30104572
- PMCID: PMC6089891
- DOI: 10.1038/s41598-018-30544-0
Determining Relative Dynamic Stability of Cell States Using Boolean Network Model
Abstract
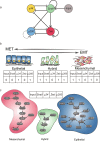
Cell state transition is at the core of biological processes in metazoan, which includes cell differentiation, epithelial-to-mesenchymal transition (EMT) and cell reprogramming. In these cases, it is important to understand the molecular mechanism of cellular stability and how the transitions happen between different cell states, which is controlled by a gene regulatory network (GRN) hard-wired in the genome. Here we use Boolean modeling of GRN to study the cell state transition of EMT and systematically compare four available methods to calculate the cellular stability of three cell states in EMT in both normal and genetically mutated cases. The results produced from four methods generally agree but do not totally agree with each other. We show that distribution of one-degree neighborhood of cell states, which are the nearest states by Hamming distance, causes the difference among the methods. From that, we propose a new method based on one-degree neighborhood, which is the simplest one and agrees with other methods to estimate the cellular stability in all scenarios of our EMT model. This new method will help the researchers in the field of cell differentiation and cell reprogramming to calculate cellular stability using Boolean model, and then rationally design their experimental protocols to manipulate the cell state transition.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

