Pan-cancer deconvolution of tumour composition using DNA methylation
- PMID: 30104673
- PMCID: PMC6089972
- DOI: 10.1038/s41467-018-05570-1
Pan-cancer deconvolution of tumour composition using DNA methylation
Erratum in
-
Author Correction: Pan-cancer deconvolution of tumour composition using DNA methylation.Nat Commun. 2018 Nov 2;9(1):4642. doi: 10.1038/s41467-018-07155-4. Nat Commun. 2018. PMID: 30389940 Free PMC article.
Abstract
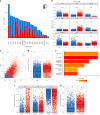
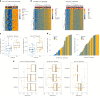
The nature and extent of immune cell infiltration into solid tumours are key determinants of therapeutic response. Here, using a DNA methylation-based approach to tumour cell fraction deconvolution, we report the integrated analysis of tumour composition and genomics across a wide spectrum of solid cancers. Initially studying head and neck squamous cell carcinoma, we identify two distinct tumour subgroups: 'immune hot' and 'immune cold', which display differing prognosis, mutation burden, cytokine signalling, cytolytic activity and oncogenic driver events. We demonstrate the existence of such tumour subgroups pan-cancer, link clonal-neoantigen burden to cytotoxic T-lymphocyte infiltration, and show that transcriptional signatures of hot tumours are selectively engaged in immunotherapy responders. We also find that treatment-naive hot tumours are markedly enriched for known immune-resistance genomic alterations, potentially explaining the heterogeneity of immunotherapy response and prognosis seen within this group. Finally, we define a catalogue of mediators of active antitumour immunity, deriving candidate biomarkers and potential targets for precision immunotherapy.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

