Turn-off colorimetric sensor for sequence-specific recognition of single-stranded DNA based upon Y-shaped DNA structure
- PMID: 30104740
- PMCID: PMC6089895
- DOI: 10.1038/s41598-018-30529-z
Turn-off colorimetric sensor for sequence-specific recognition of single-stranded DNA based upon Y-shaped DNA structure
Abstract
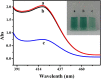
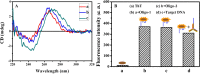
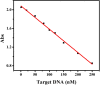
A novel turn-off colorimetric sensor for sequence-specific recognition of single-stranded DNA (ssDNA) was established by combining Y-shaped DNA duplex and G-quadruplex-hemin DNAzyme. A G-rich single-stranded DNA (Oligo-1) displays peroxidase mimicking catalytic activity due to the specific binding with hemin in the presence of K+, which was able to catalyze the oxidation of colorless 2,2'-azinobis(3-ethylbenzothiazoline)-6-sulfonic acid (ABTS2-) by H2O2 to generate green ABTS•- radical for colorimetric assay. Oligonucleotide 2 (Oligo-2) was partly complementary with Oligo-1 and the target DNA. Upon addition of target DNA, Oligo-1, Oligo-2 and target DNA can hybridize with each other to form Y-shaped DNA duplex. The DNAzyme sequence of Oligo-1 was partly caged into Y-shaped DNA duplex, resulting in the inactivation of the DNAzyme and a sharp decrease of the absorbance of the oxidation product of ABTS2-. Under the optimum condition, the absorbance decreased linearly with the concentration of target DNA over the range of 1.0-250 nM and the detection limit was 0.95 nM (3σ/slope) Moreover, satisfied result was obtained for the discrimination of single-base or two-base mismatched DNA.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

