Automated Gleason grading of prostate cancer tissue microarrays via deep learning
- PMID: 30104757
- PMCID: PMC6089889
- DOI: 10.1038/s41598-018-30535-1
Automated Gleason grading of prostate cancer tissue microarrays via deep learning
Erratum in
-
Author Correction: Automated Gleason grading of prostate cancer tissue microarrays via deep learning.Sci Rep. 2019 May 16;9(1):7668. doi: 10.1038/s41598-019-43989-8. Sci Rep. 2019. PMID: 31092857 Free PMC article.
-
Author Correction: Automated Gleason grading of prostate cancer tissue microarrays via deep learning.Sci Rep. 2021 Nov 23;11(1):23032. doi: 10.1038/s41598-021-02195-1. Sci Rep. 2021. PMID: 34815456 Free PMC article. No abstract available.
Abstract
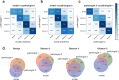
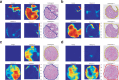
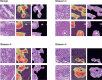
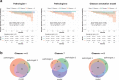
The Gleason grading system remains the most powerful prognostic predictor for patients with prostate cancer since the 1960s. Its application requires highly-trained pathologists, is tedious and yet suffers from limited inter-pathologist reproducibility, especially for the intermediate Gleason score 7. Automated annotation procedures constitute a viable solution to remedy these limitations. In this study, we present a deep learning approach for automated Gleason grading of prostate cancer tissue microarrays with Hematoxylin and Eosin (H&E) staining. Our system was trained using detailed Gleason annotations on a discovery cohort of 641 patients and was then evaluated on an independent test cohort of 245 patients annotated by two pathologists. On the test cohort, the inter-annotator agreements between the model and each pathologist, quantified via Cohen's quadratic kappa statistic, were 0.75 and 0.71 respectively, comparable with the inter-pathologist agreement (kappa = 0.71). Furthermore, the model's Gleason score assignments achieved pathology expert-level stratification of patients into prognostically distinct groups, on the basis of disease-specific survival data available for the test cohort. Overall, our study shows promising results regarding the applicability of deep learning-based solutions towards more objective and reproducible prostate cancer grading, especially for cases with heterogeneous Gleason patterns.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- WHO Classification of Tumours of the Urinary System and Male Genital Organs. International Agency for Research on Cancer (IARC) (2016).
-
- Gleason, D. F. & Mellinger, G. T. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J. Urol. 111, 58–64 (1974). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

