Investigating Effects of Tulathromycin Metaphylaxis on the Fecal Resistome and Microbiome of Commercial Feedlot Cattle Early in the Feeding Period
- PMID: 30105011
- PMCID: PMC6077226
- DOI: 10.3389/fmicb.2018.01715
Investigating Effects of Tulathromycin Metaphylaxis on the Fecal Resistome and Microbiome of Commercial Feedlot Cattle Early in the Feeding Period
Abstract
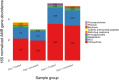
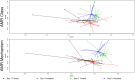
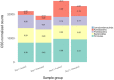
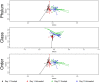
The objective was to examine effects of treating commercial beef feedlot cattle with therapeutic doses of tulathromycin, a macrolide antimicrobial drug, on changes in the fecal resistome and microbiome using shotgun metagenomic sequencing. Two pens of cattle were used, with all cattle in one pen receiving metaphylaxis treatment (800 mg subcutaneous tulathromycin) at arrival to the feedlot, and all cattle in the other pen remaining unexposed to parenteral antibiotics throughout the study period. Fecal samples were collected from 15 selected cattle in each group just prior to treatment (Day 1), and again 11 days later (Day 11). Shotgun sequencing was performed on isolated metagenomic DNA, and reads were aligned to a resistance and a taxonomic database to identify alignments to antimicrobial resistance (AMR) gene accessions and microbiome content. Overall, we identified AMR genes accessions encompassing 9 classes of AMR drugs and encoding 24 unique AMR mechanisms. Statistical analysis was used to identify differences in the resistome and microbiome between the untreated and treated groups at both timepoints, as well as over time. Based on composition and ordination analyses, the resistome and microbiome were not significantly different between the two groups on Day 1 or on Day 11. However, both the resistome and microbiome changed significantly between these two sampling dates. These results indicate that the transition into the feedlot-and associated changes in diet, geography, conspecific exposure, and environment-may exert a greater influence over the fecal resistome and microbiome of feedlot cattle than common metaphylactic antimicrobial drug treatment.
Keywords: feedlot; metagenomics; metaphylaxis; microbiome; resistome; tulathromycin.
Figures



References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed January 31, 2018).
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

