Genome-Wide Signatures of Selection Reveal Genes Associated With Performance in American Quarter Horse Subpopulations
- PMID: 30105047
- PMCID: PMC6060370
- DOI: 10.3389/fgene.2018.00249
Genome-Wide Signatures of Selection Reveal Genes Associated With Performance in American Quarter Horse Subpopulations
Abstract
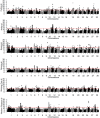
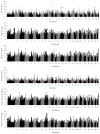
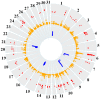
Selective breeding for athletic performance in various disciplines has resulted in population stratification within the American Quarter Horse (QH) breed. The goals of this study were to utilize high density genotype data to: (1) identify genomic regions undergoing positive selection within and among QH subpopulations; (2) investigate haplotype structure within each QH subpopulation; and (3) identify candidate genes within genomic regions of interest (ROI), as well as biological pathways, predicted to play a role in elite performance in each group. For that, 65K SNP genotyping data on 143 elite individuals from 6 QH subpopulations (cutting, halter, racing, reining, western pleasure, and working cow) were imputed to 2M SNPs. Signatures of selection were identified using FST-based (di ) and haplotype-based (hapFLK) analyses, accompanied by identification of local haplotype structure and sharing within subpopulations (hapQTL). Regions undergoing positive selection were identified on all 31 autosomes, and ROI on 2 chromosomes were identified by all 3 methods combined. Genes within each ROI were retrieved and used to identify pathways and genes that might contribute to performance in each subpopulation. These included, among others, candidate genes associated with skeletal muscle development, metabolism, and central nervous system development. This work improves our understanding of equine breed development, and provides breeders with a better understanding of how selective breeding impacts the performance of QH populations.
Keywords: SNP; ancestral haplotypes; genotyping; horse; imputation; selection signatures.
Figures
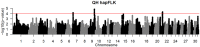
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

