A patient-derived iPSC model revealed oxidative stress increases facioscapulohumeral muscular dystrophy-causative DUX4
- PMID: 30107443
- PMCID: PMC6240734
- DOI: 10.1093/hmg/ddy293
A patient-derived iPSC model revealed oxidative stress increases facioscapulohumeral muscular dystrophy-causative DUX4
Abstract
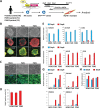
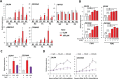
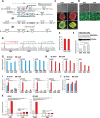
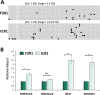
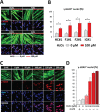
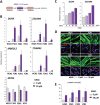
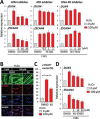
Double homeobox 4 (DUX4), the causative gene of facioscapulohumeral muscular dystrophy (FSHD), is ectopically expressed in the skeletal muscle cells of FSHD patients because of chromatin relaxation at 4q35. The diminished heterochromatic state at 4q35 is caused by either large genome contractions [FSHD type 1 (FSHD1)] or mutations in genes encoding chromatin regulators, such as SMCHD1 [FSHD type 2 (FSHD2)]. However, the mechanism by which DUX4 expression is regulated remains largely unknown. Here, using a myocyte model developed from patient-derived induced pluripotent stem cells, we determined that DUX4 expression was increased by oxidative stress (OS), a common environmental stress in skeletal muscle, in both FSHD1 and FSHD2 myocytes. We generated FSHD2-derived isogenic control clones with SMCHD1 mutation corrected by clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR associated 9 (Cas9) and homologous recombination and found in the myocytes obtained from these clones that DUX4 basal expression and the OS-induced upregulation were markedly suppressed due to an increase in the heterochromatic state at 4q35. We further found that DNA damage response (DDR) was involved in OS-induced DUX4 increase and identified ataxia-telangiectasia mutated, a DDR regulator, as a mediator of this effect. Our results suggest that the relaxed chromatin state in FSHD muscle cells permits aberrant access of OS-induced DDR signaling, thus increasing DUX4 expression. These results suggest OS could represent an environmental risk factor that promotes FSHD progression.
Figures
Similar articles
-
Intronic SMCHD1 variants in FSHD: testing the potential for CRISPR-Cas9 genome editing.J Med Genet. 2019 Dec;56(12):828-837. doi: 10.1136/jmedgenet-2019-106402. Epub 2019 Nov 1. J Med Genet. 2019. PMID: 31676591 Free PMC article.
-
The Genetics and Epigenetics of Facioscapulohumeral Muscular Dystrophy.Annu Rev Genomics Hum Genet. 2019 Aug 31;20:265-291. doi: 10.1146/annurev-genom-083118-014933. Epub 2019 Apr 24. Annu Rev Genomics Hum Genet. 2019. PMID: 31018108 Review.
-
A complex interplay of genetic and epigenetic events leads to abnormal expression of the DUX4 gene in facioscapulohumeral muscular dystrophy.Neuromuscul Disord. 2016 Dec;26(12):844-852. doi: 10.1016/j.nmd.2016.09.015. Epub 2016 Sep 19. Neuromuscul Disord. 2016. PMID: 27816329 Review.
-
Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene.PLoS Genet. 2010 Oct 28;6(10):e1001181. doi: 10.1371/journal.pgen.1001181. PLoS Genet. 2010. PMID: 21060811 Free PMC article.
-
Homozygous nonsense variant in LRIF1 associated with facioscapulohumeral muscular dystrophy.Neurology. 2020 Jun 9;94(23):e2441-e2447. doi: 10.1212/WNL.0000000000009617. Epub 2020 May 28. Neurology. 2020. PMID: 32467133 Free PMC article.
Cited by
-
Possible application of muscle specific conditional mouse-derived induced pluripotent stem cells for muscle research.Biochem Biophys Rep. 2020 Feb 1;21:100744. doi: 10.1016/j.bbrep.2020.100744. eCollection 2020 Mar. Biochem Biophys Rep. 2020. PMID: 32025579 Free PMC article.
-
Early and non-destructive prediction of the differentiation efficiency of human induced pluripotent stem cells using imaging and machine learning.Sci Rep. 2025 Jul 23;15(1):26821. doi: 10.1038/s41598-025-11108-5. Sci Rep. 2025. PMID: 40702033 Free PMC article.
-
Promising Perspective to Facioscapulohumeral Muscular Dystrophy Treatment: Nutraceuticals and Phytochemicals.Curr Neuropharmacol. 2021;19(12):2276-2295. doi: 10.2174/1570159X19666210726151924. Curr Neuropharmacol. 2021. PMID: 34315378 Free PMC article. Review.
-
p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models.Nat Genet. 2021 Aug;53(8):1207-1220. doi: 10.1038/s41588-021-00893-0. Epub 2021 Jul 15. Nat Genet. 2021. PMID: 34267371 Free PMC article.
-
Analysis of genes regulated by DUX4 via oxidative stress reveals potential therapeutic targets for treatment of facioscapulohumeral dystrophy.Redox Biol. 2021 Jul;43:102008. doi: 10.1016/j.redox.2021.102008. Epub 2021 May 13. Redox Biol. 2021. PMID: 34030118 Free PMC article.
References
-
- Zeng W., De Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A. et al. (2009) Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet., 5, e1000559. - PMC - PubMed
-
- Lemmers R.J.L.F., Goeman J.J., Van der Vliet P.J., Van Nieuwenhuizen M.P., Balog J., Vos-Versteeg M., Camano P., Ramos Arroyo M.A., Jerico I., Rogers M.T. et al. (2015) Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet., 24, 659–669. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

