High genetic diversity in hard ticks from a China-Myanmar border county
- PMID: 30107820
- PMCID: PMC6092835
- DOI: 10.1186/s13071-018-3048-5
High genetic diversity in hard ticks from a China-Myanmar border county
Abstract
Background: Many tick species have great morphological similarity and are thus grouped into species complexes. Molecular methods are therefore useful in the classification and identification of ticks. However, little is known about the genetic diversity of hard ticks in China, especially at the subspecies level. Tengchong is one of the epidemic foci of tick-borne diseases in China, but the tick species inhabiting the local area are still unknown.
Methods: Eighteen villages in Tengchong County, China, were selected for sampling carried out from September to October 2014. Infesting hard ticks were removed from the body surface of domestic animals and questing ticks were collected from grazing fields. After morphological identification, molecular characteristics of each tick species were analyzed based on both 16S rRNA and cytochrome c oxidase subunit 1 (cox1) gene fragments.
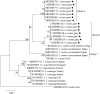
Results: Six tick species were identified based on morphology: Rhipicephalus microplus, R. haemaphysaloides, Ixodes ovatus, Haemaphysalis longicornis, H. shimoga and H. kitaokai. Phylogenetic analysis using the cox1 gene revealed that R. microplus ticks from the present study belong to clade C. For tick samples of both R. haemaphysaloides and I. ovatus, three phylogenetic groups were recognized, and the intergroup genetic distances exceeded the usual tick species boundaries. Haemaphysalis longicornis ticks were clustered into two separate clades based on the cox1 gene. For ticks from both H. shimoga and H. kitaokai, two phylogenetic groups were recognized based on the phylogenetic analysis of the 16S rRNA gene, and the intergroup genetic distances also exceeded the known boundaries for closely related tick species.
Conclusions: According to molecular analyses, new species or subspecies closely related to R. haemaphysaloides, I. ovatus, H. shimoga and H. kitaokai probably exist in the China-Myanmar border Tengchong County, or these ticks form species complexes with highly divergent mitochondrial lineages. Morphological comparisons are warranted to further confirm the taxonomic status of these tick species.
Keywords: 16S rRNA gene; China; Genetic diversity; Hard tick; Tengchong County; cox1 gene.
Conflict of interest statement
Ethics approval and consent to participate
No specific permits were required for this study. The study did not involve endangered or protected species. Therefore, the local ethics committee deemed that approval was unnecessary.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Jongejan F, Uilenberg G. The global importance of ticks. Parasitology. 2004;129(Suppl.):S3–S14. - PubMed
MeSH terms
Substances
Grants and funding
- Nos. 2016YFC1202001/National Key Research and Development Program of China
- 2016YFC1200500/National Key Research and Development Program of China
- No. 201202019/the Special Fund for Health Research in the Public Interest China
- No. 2017BSQD52/Scientific Research Foundation for Doctors of Weifang Medical College
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

