CRISPR-SKIP: programmable gene splicing with single base editors
- PMID: 30107853
- PMCID: PMC6092781
- DOI: 10.1186/s13059-018-1482-5
CRISPR-SKIP: programmable gene splicing with single base editors
Abstract
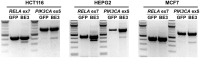
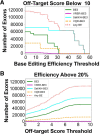
CRISPR gene editing has revolutionized biomedicine and biotechnology by providing a simple means to engineer genes through targeted double-strand breaks in the genomic DNA of living cells. However, given the stochasticity of cellular DNA repair mechanisms and the potential for off-target mutations, technologies capable of introducing targeted changes with increased precision, such as single-base editors, are preferred. We present a versatile method termed CRISPR-SKIP that utilizes cytidine deaminase single-base editors to program exon skipping by mutating target DNA bases within splice acceptor sites. Given its simplicity and precision, CRISPR-SKIP will be broadly applicable in gene therapy and synthetic biology.
Keywords: Alternative splicing; BRCA2; Base editing; CRISPR-Cas9; Exon skipping; Gene editing; Gene isoform; PIK3CA; RELA; Synthetic biology.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

