Oligodendrocyte precursor survival and differentiation requires chromatin remodeling by Chd7 and Chd8
- PMID: 30108144
- PMCID: PMC6126750
- DOI: 10.1073/pnas.1802620115
Oligodendrocyte precursor survival and differentiation requires chromatin remodeling by Chd7 and Chd8
Abstract
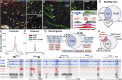
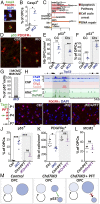
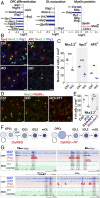
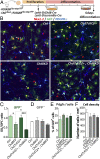
Oligodendrocyte precursor cells (OPCs) constitute the main proliferative cells in the adult brain, and deregulation of OPC proliferation-differentiation balance results in either glioma formation or defective adaptive (re)myelination. OPC differentiation requires significant genetic reprogramming, implicating chromatin remodeling. Mounting evidence indicates that chromatin remodelers play important roles during normal development and their mutations are associated with neurodevelopmental defects, with CHD7 haploinsuficiency being the cause of CHARGE syndrome and CHD8 being one of the strongest autism spectrum disorder (ASD) high-risk-associated genes. Herein, we report on uncharacterized functions of the chromatin remodelers Chd7 and Chd8 in OPCs. Their OPC-chromatin binding profile, combined with transcriptome and chromatin accessibility analyses of Chd7-deleted OPCs, demonstrates that Chd7 protects nonproliferative OPCs from apoptosis by chromatin closing and transcriptional repression of p53 Furthermore, Chd7 controls OPC differentiation through chromatin opening and transcriptional activation of key regulators, including Sox10, Nkx2.2, and Gpr17 However, Chd7 is dispensable for oligodendrocyte stage progression, consistent with Chd8 compensatory function, as suggested by their common chromatin-binding profiles and genetic interaction. Finally, CHD7 and CHD8 bind in OPCs to a majority of ASD risk-associated genes, suggesting an implication of oligodendrocyte lineage cells in ASD neurological defects. Our results thus offer new avenues to understand and modulate the CHD7 and CHD8 functions in normal development and disease.
Keywords: CHARGE; autism spectrum disorder; chromatin remodeling; oligodendrocyte; transcription regulation.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Chd7 Collaborates with Sox2 to Regulate Activation of Oligodendrocyte Precursor Cells after Spinal Cord Injury.J Neurosci. 2017 Oct 25;37(43):10290-10309. doi: 10.1523/JNEUROSCI.1109-17.2017. Epub 2017 Sep 20. J Neurosci. 2017. PMID: 28931573 Free PMC article.
-
Dual Requirement of CHD8 for Chromatin Landscape Establishment and Histone Methyltransferase Recruitment to Promote CNS Myelination and Repair.Dev Cell. 2018 Jun 18;45(6):753-768.e8. doi: 10.1016/j.devcel.2018.05.022. Dev Cell. 2018. PMID: 29920279 Free PMC article.
-
Oligodendrocyte dysfunction due to Chd8 mutation gives rise to behavioral deficits in mice.Hum Mol Genet. 2020 May 28;29(8):1274-1291. doi: 10.1093/hmg/ddaa036. Hum Mol Genet. 2020. PMID: 32142125
-
Functional Insights into Chromatin Remodelling from Studies on CHARGE Syndrome.Trends Genet. 2015 Oct;31(10):600-611. doi: 10.1016/j.tig.2015.05.009. Epub 2015 Sep 24. Trends Genet. 2015. PMID: 26411921 Free PMC article. Review.
-
Chromodomain helicase DNA-binding proteins in stem cells and human developmental diseases.Stem Cells Dev. 2015 Apr 15;24(8):917-26. doi: 10.1089/scd.2014.0544. Epub 2015 Feb 25. Stem Cells Dev. 2015. PMID: 25567374 Free PMC article. Review.
Cited by
-
Semaphorin Regulation by the Chromatin Remodeler CHD7: An Emerging Genetic Interaction Shaping Neural Cells and Neural Crest in Development and Cancer.Front Cell Dev Biol. 2021 Apr 1;9:638674. doi: 10.3389/fcell.2021.638674. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869187 Free PMC article. Review.
-
Promyelinating drugs promote functional recovery in an autism spectrum disorder mouse model of Pitt-Hopkins syndrome.Brain. 2023 Aug 1;146(8):3331-3346. doi: 10.1093/brain/awad057. Brain. 2023. PMID: 37068912 Free PMC article.
-
Chromatin remodeller CHD7 is required for GABAergic neuron development by promoting PAQR3 expression.EMBO Rep. 2021 Jun 4;22(6):e50958. doi: 10.15252/embr.202050958. Epub 2021 Apr 26. EMBO Rep. 2021. PMID: 33900016 Free PMC article.
-
Intersection of Brain Development and Paediatric Diffuse Midline Gliomas: Potential Role of Microenvironment in Tumour Growth.Brain Sci. 2018 Nov 16;8(11):200. doi: 10.3390/brainsci8110200. Brain Sci. 2018. PMID: 30453529 Free PMC article. Review.
-
CHD8 mutations increase gliogenesis to enlarge brain size in the nonhuman primate.Cell Discov. 2023 Mar 7;9(1):27. doi: 10.1038/s41421-023-00525-3. Cell Discov. 2023. PMID: 36878905 Free PMC article.
References
-
- Tekki-Kessaris N, et al. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

