Human proteins that interact with RNA/DNA hybrids
- PMID: 30108179
- PMCID: PMC6120628
- DOI: 10.1101/gr.237362.118
Human proteins that interact with RNA/DNA hybrids
Abstract
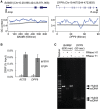
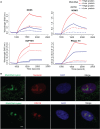
RNA/DNA hybrids form when RNA hybridizes with its template DNA generating a three-stranded structure known as the R-loop. Knowledge of how they form and resolve, as well as their functional roles, is limited. Here, by pull-down assays followed by mass spectrometry, we identified 803 proteins that bind to RNA/DNA hybrids. Because these proteins were identified using in vitro assays, we confirmed that they bind to R-loops in vivo. They include proteins that are involved in a variety of functions, including most steps of RNA processing. The proteins are enriched for K homology (KH) and helicase domains. Among them, more than 300 proteins preferred binding to hybrids than double-stranded DNA. These proteins serve as starting points for mechanistic studies to elucidate what RNA/DNA hybrids regulate and how they are regulated.
© 2018 Wang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Abdouni HS, King JJ, Ghorbani A, Fifield H, Berghuis L, Larijani M. 2018. DNA/RNA hybrid substrates modulate the catalytic activity of purified AID. Mol Immunol 93: 94–106. - PubMed
-
- Antanaviciute A, Baquero-Perez B, Watson CM, Harrison SM, Lascelles C, Crinnion L, Markham AF, Bonthron DT, Whitehouse A, Carr IM. 2017. m6aViewer: software for the detection, analysis, and visualization of N6-methyladenosine peaks from m6A-seq/ME-RIP sequencing data. RNA 23: 1493–1501. - PMC - PubMed
-
- Baker TA, Kornberg A. 1988. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell 55: 113–123. - PubMed
-
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. - PubMed
-
- Benevides JM, Wang AH, Rich A, Kyogoku Y, van der Marel GA, van Boom JH, Thomas GJ. 1986. Raman spectra of single crystals of r(GCG)d(CGC) and d(CCCCGGGG) as models for A DNA, their structure transitions in aqueous solution, and comparison with double-helical poly(dG)·poly(dC). Biochemistry (Mosc) 25: 41–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
