Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins
- PMID: 30108412
- PMCID: PMC6080641
- DOI: 10.1016/j.simyco.2018.06.001
Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins
Abstract
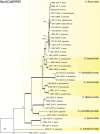
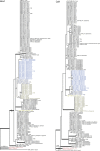
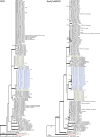
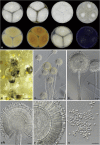
Aflatoxins and ochratoxins are among the most important mycotoxins of all and producers of both types of mycotoxins are present in Aspergillus section Flavi, albeit never in the same species. Some of the most efficient producers of aflatoxins and ochratoxins have not been described yet. Using a polyphasic approach combining phenotype, physiology, sequence and extrolite data, we describe here eight new species in section Flavi. Phylogenetically, section Flavi is split in eight clades and the section currently contains 33 species. Two species only produce aflatoxin B1 and B2 (A. pseudotamarii and A. togoensis), and 14 species are able to produce aflatoxin B1, B2, G1 and G2: three newly described species A. aflatoxiformans, A. austwickii and A. cerealis in addition to A. arachidicola, A. minisclerotigenes, A. mottae, A. luteovirescens (formerly A. bombycis), A. nomius, A. novoparasiticus, A. parasiticus, A. pseudocaelatus, A. pseudonomius, A. sergii and A. transmontanensis. It is generally accepted that A. flavus is unable to produce type G aflatoxins, but here we report on Korean strains that also produce aflatoxin G1 and G2. One strain of A. bertholletius can produce the immediate aflatoxin precursor 3-O-methylsterigmatocystin, and one strain of Aspergillus sojae and two strains of Aspergillus alliaceus produced versicolorins. Strains of the domesticated forms of A. flavus and A. parasiticus, A. oryzae and A. sojae, respectively, lost their ability to produce aflatoxins, and from the remaining phylogenetically closely related species (belonging to the A. flavus-, A. tamarii-, A. bertholletius- and A. nomius-clades), only A. caelatus, A. subflavus and A. tamarii are unable to produce aflatoxins. With exception of A. togoensis in the A. coremiiformis-clade, all species in the phylogenetically more distant clades (A. alliaceus-, A. coremiiformis-, A. leporis- and A. avenaceus-clade) are unable to produce aflatoxins. Three out of the four species in the A. alliaceus-clade can produce the mycotoxin ochratoxin A: A. alliaceus s. str. and two new species described here as A. neoalliaceus and A. vandermerwei. Eight species produced the mycotoxin tenuazonic acid: A. bertholletius, A. caelatus, A. luteovirescens, A. nomius, A. pseudocaelatus, A. pseudonomius, A. pseudotamarii and A. tamarii while the related mycotoxin cyclopiazonic acid was produced by 13 species: A. aflatoxiformans, A. austwickii, A. bertholletius, A. cerealis, A. flavus, A. minisclerotigenes, A. mottae, A. oryzae, A. pipericola, A. pseudocaelatus, A. pseudotamarii, A. sergii and A. tamarii. Furthermore, A. hancockii produced speradine A, a compound related to cyclopiazonic acid. Selected A. aflatoxiformans, A. austwickii, A. cerealis, A. flavus, A. minisclerotigenes, A. pipericola and A. sergii strains produced small sclerotia containing the mycotoxin aflatrem. Kojic acid has been found in all species in section Flavi, except A. avenaceus and A. coremiiformis. Only six species in the section did not produce any known mycotoxins: A. aspearensis, A. coremiiformis, A. lanosus, A. leporis, A. sojae and A. subflavus. An overview of other small molecule extrolites produced in Aspergillus section Flavi is given.
Keywords: A. Nováková; A. vandermerwei Frisvad; Aflatoxins; Arzanlou & Samson; Aspergillus; Aspergillus aflatoxiformans Frisvad; Aspergillus aspearensis Houbraken; Aspergillus austwickii Frisvad; Aspergillus cerealis Houbraken; Aspergillus neoalliaceus A. Nováková; Aspergillus pipericola Frisvad; Aspergillus subflavus Hubka; Cyclopiazonic acid; Ezekiel; Ezekiel & Samson; Frisvad; Frisvad & Houbraken; Hubka; Samson; Samson & Houbraken; Section Flavi; Tenuazonic acid.
Figures

















References
-
- Adler M., Wintersteiner O. A reinvestigation of flavacidin, the penicillin produced by Aspergillus flavus. Journal of Biological Chemistry. 1948;176:873–891. - PubMed
-
- Amaike S., Keller N.P. Aspergillus flavus. Annual Review of Phytopathology. 2011;49:107–133. - PubMed
-
- Amare M.G., Keller N.P. Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genetics and Biology. 2014;66:11–18. - PubMed
-
- Ammar H.A.M., Srour A.Y., Ezzat S.M. Identification and characterization of genes involved in kojic acid biosynthesis in Aspergillus flavus. Annals of Microbiology. 2017;67:691–702.
LinkOut - more resources
Full Text Sources
Other Literature Sources
