Do Ruminal Ciliates Select Their Preys and Prokaryotic Symbionts?
- PMID: 30108566
- PMCID: PMC6079354
- DOI: 10.3389/fmicb.2018.01710
Do Ruminal Ciliates Select Their Preys and Prokaryotic Symbionts?
Abstract
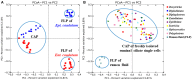
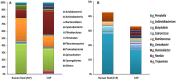
Ruminal ciliates both preys on and form symbiotic relationships with other members of the ruminal microbiota for their survival. However, it remains elusive if they have selectivity over their preys or symbionts. In the present study, we investigated the above selectivity by identifying and comparing the free-living prokaryotes (FLP) and the ciliate-associated prokaryotes (CAP) using Illumina MiSeq sequencing of 16S rRNA gene amplicons. We used single ciliates cells of both monocultures of Entodinium caudatum and Epidinium caudatum and eight different ciliate genera isolated from fresh rumen fluid of dairy cows. Irrespective of the source (laboratory monocultures vs. fresh isolates) of the single ciliate cells, the CAP significantly differed from the FLP in microbiota community profiles. Many bacterial taxa were either enriched or almost exclusively found in the CAP across most of the ciliate genera. A number of bacteria were also found for the first time as ruminal bacteria in the CAP. However, no clear difference was found in methanogens between the CAP and the FLP, which was confirmed using methanogen-specific qPCR. These results suggest that ruminal ciliates probably select their preys and symbionts, the latter of which has rarely been found among the free-living ruminal prokaryotes. The bacteria enriched or exclusively found in the CAP can be target bacteria to detect and localize using specific probes designed from their 16S rRNA sequences, to characterize using single-cell genomics, or to isolate using new media designed based on genomic information.
Keywords: ciliate-associated prokaryotes; free-living prokaryotes; preys; ruminal ciliates; symbionts.
Figures



Similar articles
-
The transcriptome of the rumen ciliate Entodinium caudatum reveals some of its metabolic features.BMC Genomics. 2019 Dec 21;20(1):1008. doi: 10.1186/s12864-019-6382-x. BMC Genomics. 2019. PMID: 31864285 Free PMC article.
-
Effect of the rumen ciliates Entodinium caudatum, Epidinium ecaudatum and Eudiplodinium maggii, and combinations thereof, on ruminal fermentation and total tract digestion in sheep.Arch Anim Nutr. 2012 Jun;66(3):180-99. doi: 10.1080/1745039x.2012.676817. Arch Anim Nutr. 2012. PMID: 22724165
-
Inhibition of the Rumen Ciliate Entodinium caudatum by Antibiotics.Front Microbiol. 2017 Jun 28;8:1189. doi: 10.3389/fmicb.2017.01189. eCollection 2017. Front Microbiol. 2017. PMID: 28702015 Free PMC article.
-
Symbionts of Ciliates and Ciliates as Symbionts.Indian J Microbiol. 2024 Jun;64(2):304-317. doi: 10.1007/s12088-024-01203-y. Epub 2024 Feb 10. Indian J Microbiol. 2024. PMID: 39010998 Free PMC article. Review.
-
Extending Burk Dehority's Perspectives on the Role of Ciliate Protozoa in the Rumen.Front Microbiol. 2020 Feb 28;11:123. doi: 10.3389/fmicb.2020.00123. eCollection 2020. Front Microbiol. 2020. PMID: 32184759 Free PMC article. Review.
Cited by
-
The transcriptome of the rumen ciliate Entodinium caudatum reveals some of its metabolic features.BMC Genomics. 2019 Dec 21;20(1):1008. doi: 10.1186/s12864-019-6382-x. BMC Genomics. 2019. PMID: 31864285 Free PMC article.
-
Pre-weaning Ruminal Administration of Differentially-Enriched, Rumen-Derived Inocula Shaped Rumen Bacterial Communities and Co-occurrence Networks of Post-weaned Dairy Calves.Front Microbiol. 2021 Feb 26;12:625488. doi: 10.3389/fmicb.2021.625488. eCollection 2021. Front Microbiol. 2021. PMID: 33717013 Free PMC article.
-
- Invited Review - Ruminal ciliates as modulators of the rumen microbiome.Anim Biosci. 2024 Feb;37(2):385-395. doi: 10.5713/ab.23.0309. Epub 2023 Dec 29. Anim Biosci. 2024. PMID: 38186255 Free PMC article.
-
Methane emission, nitrogen excretion, and energy partitioning in Hanwoo steers fed a typical TMR diet supplemented with Pharbitis nil seeds.Front Vet Sci. 2024 Sep 24;11:1467077. doi: 10.3389/fvets.2024.1467077. eCollection 2024. Front Vet Sci. 2024. PMID: 39380775 Free PMC article.
-
Repeated Inoculation of Young Calves With Rumen Microbiota Does Not Significantly Modulate the Rumen Prokaryotic Microbiota Consistently but Decreases Diarrhea.Front Microbiol. 2020 Jun 24;11:1403. doi: 10.3389/fmicb.2020.01403. eCollection 2020. Front Microbiol. 2020. PMID: 32670244 Free PMC article.
References
-
- Amaral-Zettler L., Artigas L. F., Baross J., Bharathi L., Boetius A., Chandramohan D., et al. (2010). A global census of marine microbes, in Life in the World's Oceans: Diversity, Distribution and Abundance ed McIntyre A. (Oxford: Blackwell Publishing Ltd.), 223–245.
-
- Baraka T. (2012). Comparative studies of rumen PH, total protozoa count, generic and species composition of ciliates in camel, buffalo, cattle, sheep and goat in Egypt. J. Am. Sci. 8, 655–669. 10.5564/mjas.v19i3.730 - DOI
-
- Bonhomme A., Durand M., Quintana C., Halpern S. (1982a). Influence du cobalt et de la vitamine B12 sur la croissance et la survie des ciliés du rumen in vitro, en fonction de la population bactérienne. Reprod. Nutr. Dévelop. 22, 107–122. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

