Identification and Discrimination of Salmonella enterica Serovar Gallinarum Biovars Pullorum and Gallinarum Based on a One-Step Multiplex PCR Assay
- PMID: 30108571
- PMCID: PMC6079294
- DOI: 10.3389/fmicb.2018.01718
Identification and Discrimination of Salmonella enterica Serovar Gallinarum Biovars Pullorum and Gallinarum Based on a One-Step Multiplex PCR Assay
Abstract
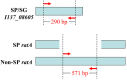
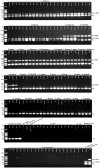
Salmonella enterica serovar Gallinarum biovars Pullorum (S. Pullorum) and Gallinarum (S. Gallinarum) can result in pullorum disease and fowl typhoid in avian species, respectively, and cause considerable economic losses in poultry in many developing countries. Conventional Salmonella serotyping is a time-consuming, labor-intensive and expensive process, and the two biovars cannot be distinguished using the traditional serological method. In this study, we developed a rapid and reliable one-step multiplex polymerase chain reaction (PCR) assay to simultaneously identify and discriminate the biovars Pullorum and Gallinarum. The multiplex PCR method focused on three specific genes, stn, I137_08605 and ratA. Based on bioinformatics analysis, we found that gene I137_08605 was present only in S. Pullorum and S. Gallinarum, and a region of difference in ratA was deleted only in S. Pullorum after comparison with that of S. Gallinarum and other Salmonella serovars. Three pairs of primers specific for the three genes were designed for the multiplex PCR system and their selectivity and sensitivity were determined. The multiplex PCR results showed that S. Pullorum and S. Gallinarum could be identified and discriminated accurately from all tested strains including 124 strains of various Salmonella serovars and 42 strains of different non-Salmonella pathogens. In addition, this multiplex PCR assay could detect a minimum genomic DNA concentration of 67.4 pg/μL, and 100 colony forming units. The efficiency of the multiplex PCR was evaluated by detecting natural-occurring Salmonella isolates from a chicken farm. The results demonstrated that the established multiplex PCR was able to identify S. Gallinarum and S. Pullorum individually, with results being consistent with traditional serotyping and biochemical testing. These results demonstrated that a highly accurate and simple biovar-specific multiplex PCR assay could be performed for the rapid identification and discrimination of Salmonella biovars Gallinarum and Pullorum, which will be useful, particularly under massive screening situations.
Keywords: Salmonella Gallinarum; Salmonella Pullorum; accurate discrimination; multiplex PCR; one-step diagnostic PCR.
Figures
References
-
- Batista D. F., Freitas Neto O. C., Barrow P. A., Oliveira M. T., Almeida A. M., Ferraudo A. S., et al. (2015). Identification and characterization of regions of difference between the Salmonella Gallinarum biovar Gallinarum and the Salmonella Gallinarum biovar Pullorum genomes. Infect. Genet. Evol. 30 74–81. 10.1016/j.meegid.2014.12.007 - DOI - PubMed
-
- Berchieri A., Jr., Murphy C. K., Marston K., Barrow P. A. (2001). Observations on the persistence and vertical transmission of Salmonella enterica serovars Pullorum and Gallinarum in chickens: effect of bacterial and host genetic background. Avian Pathol. 30 221–231. 10.1080/03079450120054631 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

