Hypoxia inducible factor down-regulation, cancer and cancer stem cells (CSCs): ongoing success stories
- PMID: 30108689
- PMCID: PMC6071925
- DOI: 10.1039/c6md00432f
Hypoxia inducible factor down-regulation, cancer and cancer stem cells (CSCs): ongoing success stories
Abstract
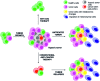
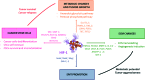
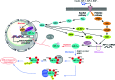
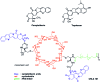
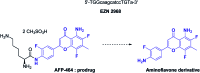
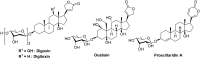
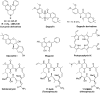
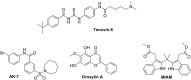
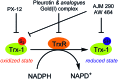
In cancers, hypoxia inducible factor 1 (HIF-1) is an over-expressed transcription factor, which regulates a large set of genes involved in tumour vascularization, metastases, and cancer stem cells (CSCs) formation and self-renewal. This protein has been identified as a relevant target in oncology and several HIF-1 modulators are now marketed or in advanced clinical trials. The purpose of this review is to summarize the advances in the understanding of its regulation and its inhibition, from the medicinal chemist point of view. To this end, we selected in the recent literature relevant examples of "hit" compounds, including small-sized organic molecules, pseudopeptides and nano-drugs, exhibiting in vitro and/or in vivo both anti-HIF-1 and anti-tumour activities. Whenever possible, a particular emphasis has been dedicated to compounds that selectively target CSCs.
Figures


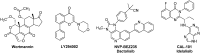


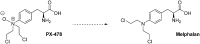
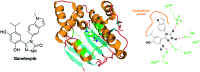









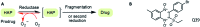
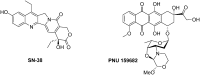




References
-
- Liu J., Liu Y., Bu W., Bu J., Sun Y., Du J., Shi J., Vaupel P., Höckel M., Mayer A. J. Am. Chem. Soc. Antioxid. Redox Signaling. 2014;2007;1369:9701. 1221. - PubMed
-
- Bhatt A. N., Chauhan A., Khanna S., Rai Y., Singh S., Soni R., Kalra N., Dwarakanath B. S., Emara M., Allalunis-Turner J., Camaj P., Carsten J., Krebs S., DeToni E. N., Blum H., Jauch K.-W., Nelson P. J., Bruns C. J., Bao B., Aamir A., Kong D., Ali S., Azmi A. S., Li Y., Banerjee S., Padhye S., Sarkar F. H., Hill R. P., Marie-Egyptienne D. T., Hedley D. W., Axelson H., Fredlund E., Ovenberger M., Landberg G., Påhlman S., Jögi A., Øra I., Nilsson H., Poellinger L., Axelson H., Påhlman S. BMC Cancer. Oncol. Rep. Mol. Cancer Res. PLoS One. Semin. Radiat. Oncol. Semin. Cell Dev. Biol. Cancer Lett. 2015;2014;2014;2012;2009;2005;2003;15311271916197:335. 1947, 421, e43726, 106, 554, 145. - PMC - PubMed
-
- Asgari Y., Zabihinpour Z., Salehzadeh-Yazdi A., Schreiber F., Masoudi-Nejad A., Chen X., Qian Y., Wu S., Zhang W., Zhang S.-L., Hu X., Tam K. Y. Genomics. Free Radical Biol. Med. Int. J. Biol. Sci. 2015;2015;2015;1057911:275. 253, 1390. - PubMed
-
- Sancho P., Barneda D., Heeschen C., Elf S., Li R., Xia S., Pan Y., Shan C., Wu S., Lonial S., Gaddh M., Arellano M. L., Khoury H. J., Khuri F. R., Lee B. H., Boggon T. J., Fan J., Chen J., Chen Y., Xu Q., Ji D., Wei Y., Chen H., Li T., Wan B., Yuan L., Huang R., Chen G. Br. J. Cancer. Oncogene. Tumour Biol. 2016;2016;2016;11437:1305. 6027. doi: 10.1038/onc.2016.196. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

