Novel T-C@AgNPs mediated biocidal mechanism against biofilm associated methicillin-resistant Staphylococcus aureus (Bap-MRSA) 090, cytotoxicity and its molecular docking studies
- PMID: 30108735
- PMCID: PMC6072311
- DOI: 10.1039/c7md00486a
Novel T-C@AgNPs mediated biocidal mechanism against biofilm associated methicillin-resistant Staphylococcus aureus (Bap-MRSA) 090, cytotoxicity and its molecular docking studies
Abstract
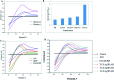
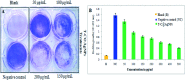
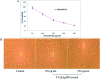
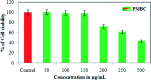
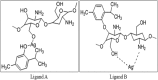
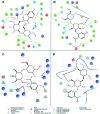
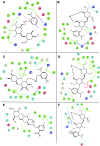
Staphylococcus aureus is a commonly found pathogen that can cause food-spoilage and life threatening infections. However, the potential molecular effects of natural active thymol molecules and chitosan silver nanoparticles (C@AgNPs) in bacteria remain unclear. This gap in the literature has prompted us to study the effects of thymol loaded chitosan silver nanoparticles (T-C@AgNPs) against biofilm associated proteins in methicillin-resistant S. aureus (Bap-MRSA) 090 and also their toxicity, anti-cancer activity, and validation of their in silico molecular docking. The results showed excellent antibacterial activity of T-C@AgNPs against Bap-MRSA 090, having a minimum inhibitory concentration of 100 μg mL-1 and a 10.08 ± 0.06 mm zone of inhibition (ZOI). The cyclic voltammogram (CV) analysis clearly showed pore forming of T-C@AgNPs at 300 μg mL-1 concentration, and evidence of the interruption of the electron transport chain was clearly seen. The 200 μg mL-1 concentration exhibited a 52.60 ± 0.25% anti-biofilm property by T-C@AgNPs against Bap-MRSA 090. The T-C@AgNPs showed no toxicity to peripheral blood mononuclear cells (PBMC) (IC50 = 221 ± 0.71 μg mL-1) compared to the control, and anti-cancer activity against human triple negative breast cancer cell line (MDA-MB-231) (IC50 110 ± 1.0 μg mL-1) compared to the standard drug Doxorubicin (IC50 = 19 ± 1.0). The excellent properties of T-C@AgNPs were validated by in silico molecular docking studies and showed best match scoring to target proteins compared to standards. These excellent properties of T-C@AgNPs highlight for the first time its pharmacology and potential in medicinal drug development applications for future research.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

