Structure-guided approach identifies a novel class of HIV-1 ribonuclease H inhibitors: binding mode insights through magnesium complexation and site-directed mutagenesis studies
- PMID: 30108947
- PMCID: PMC6072344
- DOI: 10.1039/c7md00600d
Structure-guided approach identifies a novel class of HIV-1 ribonuclease H inhibitors: binding mode insights through magnesium complexation and site-directed mutagenesis studies
Abstract
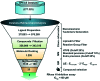
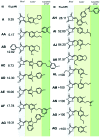
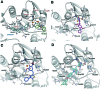
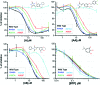
Persistent HIV infection requires lifelong treatment and among the 2.1 million new HIV infections that occur every year there is an increased rate of transmitted drug-resistant mutations. This fact requires a constant and timely effort in order to identify and develop new HIV inhibitors with innovative mechanisms. The HIV-1 reverse transcriptase (RT) associated ribonuclease H (RNase H) is the only viral encoded enzyme that still lacks an efficient inhibitor despite the fact that it is a well-validated target whose functional abrogation compromises viral infectivity. Identification of new drugs is a long and expensive process that can be speeded up by in silico methods. In the present study, a structure-guided screening is coupled with a similarity-based search on the Specs database to identify a new class of HIV-1 RNase H inhibitors. Out of the 45 compounds selected for experimental testing, 15 inhibited the RNase H function below 100 μM with three hits exhibiting IC50 values <10 μM. The most active compound, AA, inhibits HIV-1 RNase H with an IC50 of 5.1 μM and exhibits a Mg-independent mode of inhibition. Site-directed mutagenesis studies provide valuable insight into the binding mode of newly identified compounds; for instance, compound AA involves extensive interactions with a lipophilic pocket formed by Ala502, Lys503, and Trp (406, 426 and 535) and polar interactions with Arg557 and the highly conserved RNase H primer-grip residue Asn474. The structural insights obtained from this work provide the bases for further lead optimization.
Figures




Similar articles
-
Ribonuclease H/DNA Polymerase HIV-1 Reverse Transcriptase Dual Inhibitor: Mechanistic Studies on the Allosteric Mode of Action of Isatin-Based Compound RMNC6.PLoS One. 2016 Jan 22;11(1):e0147225. doi: 10.1371/journal.pone.0147225. eCollection 2016. PLoS One. 2016. PMID: 26800261 Free PMC article.
-
Targeting HIV-1 RNase H: N'-(2-Hydroxy-benzylidene)-3,4,5-Trihydroxybenzoylhydrazone as Selective Inhibitor Active against NNRTIs-Resistant Variants.Viruses. 2020 Jul 6;12(7):729. doi: 10.3390/v12070729. Viruses. 2020. PMID: 32640577 Free PMC article.
-
Cutting into the Substrate Dominance: Pharmacophore and Structure-Based Approaches toward Inhibiting Human Immunodeficiency Virus Reverse Transcriptase-Associated Ribonuclease H.Acc Chem Res. 2020 Jan 21;53(1):218-230. doi: 10.1021/acs.accounts.9b00450. Epub 2019 Dec 27. Acc Chem Res. 2020. PMID: 31880912 Free PMC article. Review.
-
Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone.Biochemistry. 1997 Mar 18;36(11):3179-85. doi: 10.1021/bi9624696. Biochemistry. 1997. PMID: 9115994
-
Small-molecule Inhibitors of HIV-1 Reverse Transcriptase-Associated Ribonuclease H Function: Challenges and Recent Developments.Curr Med Chem. 2021;28(30):6146-6178. doi: 10.2174/0929867328666210322164557. Curr Med Chem. 2021. PMID: 34225606 Review.
Cited by
-
Design, synthesis, and biologic evaluation of novel galloyl derivatives as HIV-1 RNase H inhibitors.Chem Biol Drug Des. 2019 Apr;93(4):582-589. doi: 10.1111/cbdd.13455. Epub 2019 Jan 24. Chem Biol Drug Des. 2019. PMID: 30560566 Free PMC article.
-
N-Terminus-Mediated Solution Structure of Dimerization Domain of PRC1.Curr Issues Mol Biol. 2022 Apr 10;44(4):1626-1645. doi: 10.3390/cimb44040111. Curr Issues Mol Biol. 2022. PMID: 35723369 Free PMC article.
-
2-(Arylamino)-6-(trifluoromethyl)nicotinic Acid Derivatives: New HIV-1 RT Dual Inhibitors Active on Viral Replication.Molecules. 2020 Mar 15;25(6):1338. doi: 10.3390/molecules25061338. Molecules. 2020. PMID: 32183488 Free PMC article.
-
RNase HI Depletion Strongly Potentiates Cell Killing by Rifampicin in Mycobacteria.Antimicrob Agents Chemother. 2022 Oct 18;66(10):e0209121. doi: 10.1128/aac.02091-21. Epub 2022 Sep 26. Antimicrob Agents Chemother. 2022. PMID: 36154174 Free PMC article.
-
The Journey of HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs) from Lab to Clinic.J Med Chem. 2019 May 23;62(10):4851-4883. doi: 10.1021/acs.jmedchem.8b00843. Epub 2018 Dec 27. J Med Chem. 2019. PMID: 30516990 Free PMC article.
References
-
- Fauci A. S. Nat. Med. 2003;9:839–843. - PubMed
-
- Gupta R. K., Gregson J., Parkin N., Haile-Selassie H., Tanuri A., Andrade Forero L., Kaleebu P., Watera C., Aghokeng A., Mutenda N., Dzangare J., Hone S., Hang Z. Z., Garcia J., Garcia Z., Marchorro P., Beteta E., Giron A., Hamers R., Inzaule S., Frenkel L. M., Chung M. H., de Oliveira T., Pillay D., Naidoo K., Kharsany A., Kugathasan R., Cutino T., Hunt G., Avila Rios S., Doherty M., Jordan M. R., Bertagnolio S. Lancet Infect. Dis. 2017 doi: 10.1016/S1473-3099(17)30702-8. - DOI - PubMed
-
- De Clercq E. Curr. Med. Chem. 2001;8:1543–1572. - PubMed
-
- UNAIDS report, on the global AIDS epidemic 2012: Join UN program on HIV/AIDS, 2012.
-
- De Clercq E. Biochim. Biophys. Acta, Mol. Basis Dis. 2002;1587:258–275. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

