Increased glutamine anabolism sensitizes non-small cell lung cancer to gefitinib treatment
- PMID: 30109143
- PMCID: PMC6085389
- DOI: 10.1038/s41420-018-0086-x
Increased glutamine anabolism sensitizes non-small cell lung cancer to gefitinib treatment
Erratum in
-
Erratum: Publisher Correction: articles initially published in wrong volume.Cell Death Discov. 2019 Jul 10;5:116. doi: 10.1038/s41420-019-0186-2. eCollection 2019. Cell Death Discov. 2019. PMID: 31312525 Free PMC article.
Abstract
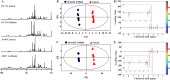
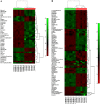
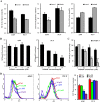
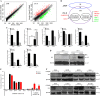
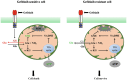
To better understand the resistance mechanism of non-small cell lung cancers (NSCLCs) to gefitinib, the metabolic profiles of gefitinib-resistant A549 cells and gefitinib-sensitive PC-9 cells were analyzed with a metabolomics analytical platform. A549 and PC-9 cells exhibited significant differences in the levels of glutamine-related metabolites. After gefitinib treatment, the glutamine level decreased in A549 cells but showed no change in PC-9 cells. The glutamine consumed by A549 cells was used to generate ATP and glutathione (GSH). As glutamine utilization was suppressed in gefitinib-treated PC-9 cells, the resulting ATP shortage and ROS accumulation led to cell death. The difference in glutamine metabolism was caused by differential changes in the levels of glutamine synthetase (GS, encoded by glutamate-ammonia ligase (GLUL)). GLUL expression was upregulated in gefitinib-sensitive cells, but it was either absent from gefitinib-resistant cells or no significant change was observed in the gefitinib-treated cells. GLUL overexpression in A549 cells significant sensitized them to gefitinib and decreased their invasive capacity. Conversely, knockout GS in PC-9 cells reduced gefitinib sensitivity and enhanced metastasis. Furthermore, the continuous exposure of gefitinib-sensitive HCC827 cells to gefitinib created gefitinib-resistant (GR) HCC827 cells, which exhibited a GLUL deletion and resistance to gefitinib. Thus, GLUL plays a vital role in determining the sensitivity of NSCLCs to gefitinib. Elevated GS levels mediate increased glutamine anabolism, and this novel mechanism sensitizes NSCLCs to gefitinib. The inhibition of glutamine utilization may serve as a potential therapeutic strategy to overcome gefitinib resistance in the clinic.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

