Retroviral nucleocapsid proteins and DNA strand transfers
- PMID: 30109196
- PMCID: PMC6088434
- DOI: 10.1016/j.biopen.2018.07.001
Retroviral nucleocapsid proteins and DNA strand transfers
Abstract
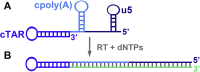
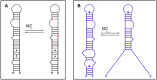
An infectious retroviral particle contains 1000-1500 molecules of the nucleocapsid protein (NC) that cover the diploid RNA genome. NC is a small zinc finger protein that possesses nucleic acid chaperone activity that enables NC to rearrange DNA and RNA molecules into the most thermodynamically stable structures usually those containing the maximum number of base pairs. Thanks to the chaperone activity, NC plays an essential role in reverse transcription of the retroviral genome by facilitating the strand transfer reactions of this process. In addition, these reactions are involved in recombination events that can generate multiple drug resistance mutations in the presence of anti-HIV-1 drugs. The strand transfer reactions rely on base pairing of folded DNA/RNA structures. The molecular mechanisms responsible for NC-mediated strand transfer reactions are presented and discussed in this review. Antiretroviral strategies targeting the NC-mediated strand transfer events are also discussed.
Keywords: HIV; Nucleocapsid protein; Retrovirus; Reverse transcription; Strand transfer.
Figures

Similar articles
-
Nucleic acid conformational changes essential for HIV-1 nucleocapsid protein-mediated inhibition of self-priming in minus-strand transfer.J Mol Biol. 2003 Jan 3;325(1):1-10. doi: 10.1016/s0022-2836(02)01177-4. J Mol Biol. 2003. PMID: 12473448
-
Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer.J Virol. 2000 Oct;74(19):8980-8. doi: 10.1128/jvi.74.19.8980-8988.2000. J Virol. 2000. PMID: 10982342 Free PMC article.
-
Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function.J Mol Biol. 2006 Nov 10;363(5):867-77. doi: 10.1016/j.jmb.2006.08.070. Epub 2006 Aug 30. J Mol Biol. 2006. PMID: 16997322
-
First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses.J Mol Biol. 1995 Dec 8;254(4):523-37. doi: 10.1006/jmbi.1995.0635. J Mol Biol. 1995. PMID: 7500330 Review.
-
Strand transfer events during HIV-1 reverse transcription.Virus Res. 2008 Jun;134(1-2):19-38. doi: 10.1016/j.virusres.2007.12.017. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279992 Review.
Cited by
-
RNA-Binding Domains of Heterologous Viral Proteins Substituted for Basic Residues in the RSV Gag NC Domain Restore Specific Packaging of Genomic RNA.Viruses. 2020 Mar 27;12(4):370. doi: 10.3390/v12040370. Viruses. 2020. PMID: 32230826 Free PMC article.
-
RNA Structural Requirements for Nucleocapsid Protein-Mediated Extended Dimer Formation.Viruses. 2022 Mar 15;14(3):606. doi: 10.3390/v14030606. Viruses. 2022. PMID: 35337013 Free PMC article.
-
Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro.Science. 2020 Oct 9;370(6513):eabc8420. doi: 10.1126/science.abc8420. Science. 2020. PMID: 33033190 Free PMC article.
-
Reverse transcription of plasma-derived HIV-1 RNA generates multiple artifacts through tRNA(Lys-3)-priming.Microbiol Spectr. 2024 Apr 2;12(4):e0387223. doi: 10.1128/spectrum.03872-23. Epub 2024 Mar 5. Microbiol Spectr. 2024. PMID: 38442427 Free PMC article.
-
The HIV-1 capsid serves as a nanoscale reaction vessel for reverse transcription.PLoS Pathog. 2024 Sep 3;20(9):e1011810. doi: 10.1371/journal.ppat.1011810. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39226318 Free PMC article.
References
-
- Laskey S.B., Siliciano R.F. A mechanistic theory to explain the efficacy of antiretroviral therapy. Nat. Rev. Microbiol. 2014;12:772–780. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

