The nuclear receptor superfamily: A structural perspective
- PMID: 30109749
- PMCID: PMC6201731
- DOI: 10.1002/pro.3496
The nuclear receptor superfamily: A structural perspective
Abstract
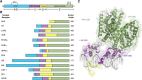
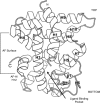
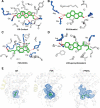
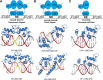
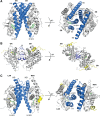
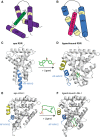
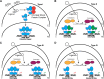
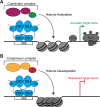
Nuclear receptors (NRs) are a family of transcription factors that regulate numerous physiological processes such as metabolism, reproduction, inflammation, as well as the circadian rhythm. NRs sense changes in lipid metabolite levels to drive differential gene expression, producing distinct physiologic effects. This is an allosteric process whereby binding a cognate ligand and specific DNA sequences drives the recruitment of diverse transcriptional co-regulators at chromatin and ultimately transactivation or transrepression of target genes. Dysregulation of NR signaling leads to various malignances, metabolic disorders, and inflammatory disease. Given their important role in physiology and ability to respond to small lipophilic ligands, NRs have emerged as valuable therapeutic targets. Here, we summarize and discuss the recent progress on understanding the complex mechanism of action of NRs, primarily from a structural perspective. Finally, we suggest future studies to improve our understanding of NR signaling and better design drugs by integrating multiple structural and biophysical approaches.
Keywords: DNA binding domain; co-regulator; ligand binding domain; nuclear receptor; transactivation; transrepression.
© 2018 The Protein Society.
Figures
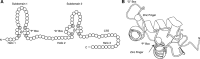
References
-
- Gustafsson JA (2016) Historical overview of nuclear receptors. J Steroid Biochem Mol Biol 157:3–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

