Accounting for non-stationarity in epidemiology by embedding time-varying parameters in stochastic models
- PMID: 30110322
- PMCID: PMC6110518
- DOI: 10.1371/journal.pcbi.1006211
Accounting for non-stationarity in epidemiology by embedding time-varying parameters in stochastic models
Erratum in
-
Correction: Accounting for non-stationarity in epidemiology by embedding time-varying parameters in stochastic models.PLoS Comput Biol. 2019 May 28;15(5):e1007062. doi: 10.1371/journal.pcbi.1007062. eCollection 2019 May. PLoS Comput Biol. 2019. PMID: 31136579 Free PMC article.
Abstract
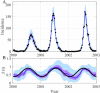
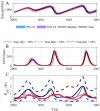
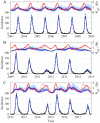
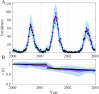
The spread of disease through human populations is complex. The characteristics of disease propagation evolve with time, as a result of a multitude of environmental and anthropic factors, this non-stationarity is a key factor in this huge complexity. In the absence of appropriate external data sources, to correctly describe the disease propagation, we explore a flexible approach, based on stochastic models for the disease dynamics, and on diffusion processes for the parameter dynamics. Using such a diffusion process has the advantage of not requiring a specific mathematical function for the parameter dynamics. Coupled with particle MCMC, this approach allows us to reconstruct the time evolution of some key parameters (average transmission rate for instance). Thus, by capturing the time-varying nature of the different mechanisms involved in disease propagation, the epidemic can be described. Firstly we demonstrate the efficiency of this methodology on a toy model, where the parameters and the observation process are known. Applied then to real datasets, our methodology is able, based solely on simple stochastic models, to reconstruct complex epidemics, such as flu or dengue, over long time periods. Hence we demonstrate that time-varying parameters can improve the accuracy of model performances, and we suggest that our methodology can be used as a first step towards a better understanding of a complex epidemic, in situation where data is limited and/or uncertain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
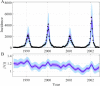
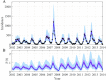
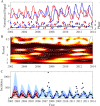
Similar articles
-
Learning stochastic process-based models of dynamical systems from knowledge and data.BMC Syst Biol. 2016 Mar 22;10:30. doi: 10.1186/s12918-016-0273-4. BMC Syst Biol. 2016. PMID: 27005698 Free PMC article.
-
Capturing the time-varying drivers of an epidemic using stochastic dynamical systems.Biostatistics. 2013 Jul;14(3):541-55. doi: 10.1093/biostatistics/kxs052. Epub 2013 Jan 4. Biostatistics. 2013. PMID: 23292757
-
Stochastic epidemic models with random environment: quasi-stationarity, extinction and final size.J Math Biol. 2013 Oct;67(4):799-831. doi: 10.1007/s00285-012-0570-5. Epub 2012 Aug 15. J Math Biol. 2013. PMID: 22892570
-
A stochastic vector-borne epidemic model: Quasi-stationarity and extinction.Math Biosci. 2017 Jul;289:89-95. doi: 10.1016/j.mbs.2017.05.004. Epub 2017 May 13. Math Biosci. 2017. PMID: 28511957
-
A statistical approach to quasi-extinction forecasting.Ecol Lett. 2007 Dec;10(12):1182-98. doi: 10.1111/j.1461-0248.2007.01105.x. Epub 2007 Sep 4. Ecol Lett. 2007. PMID: 17803676 Review.
Cited by
-
Improving Dengue Forecasts by Using Geospatial Big Data Analysis in Google Earth Engine and the Historical Dengue Information-Aided Long Short Term Memory Modeling.Biology (Basel). 2022 Jan 21;11(2):169. doi: 10.3390/biology11020169. Biology (Basel). 2022. PMID: 35205036 Free PMC article.
-
Spatially explicit effective reproduction numbers from incidence and mobility data.Proc Natl Acad Sci U S A. 2023 May 16;120(20):e2219816120. doi: 10.1073/pnas.2219816120. Epub 2023 May 9. Proc Natl Acad Sci U S A. 2023. PMID: 37159476 Free PMC article.
-
Estimation of the reproduction number of influenza A(H1N1)pdm09 in South Korea using heterogeneous models.BMC Infect Dis. 2021 Jul 7;21(1):658. doi: 10.1186/s12879-021-06121-8. BMC Infect Dis. 2021. PMID: 34233622 Free PMC article.
-
Bayesian workflow for time-varying transmission in stratified compartmental infectious disease transmission models.PLoS Comput Biol. 2024 Apr 29;20(4):e1011575. doi: 10.1371/journal.pcbi.1011575. eCollection 2024 Apr. PLoS Comput Biol. 2024. PMID: 38683878 Free PMC article.
-
Modeling the time-dependent transmission rate using gaussian pulses for analyzing the COVID-19 outbreaks in the world.Sci Rep. 2023 Mar 18;13(1):4466. doi: 10.1038/s41598-023-31714-5. Sci Rep. 2023. PMID: 36934167 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

